We selected fission yeast as our model due to the ease of genetic manipulation, its shared regulatory pathways with mammals, and the presence of two rDNA arrays as opposed to the single array in budding yeast.
Why fission yeast?
-
Like most yeast, they have genomes that are easy to manipulate. Their genome easily undergo change in response to environmental factors and they easily uptake DNA up to a whole chromosome size. There also exists an extensive genetic engineering toolbox for yeast
-
Yeast and mammals share a lot of homology because they share a common ancestor. There are many cellular pathways that are common to both.
-
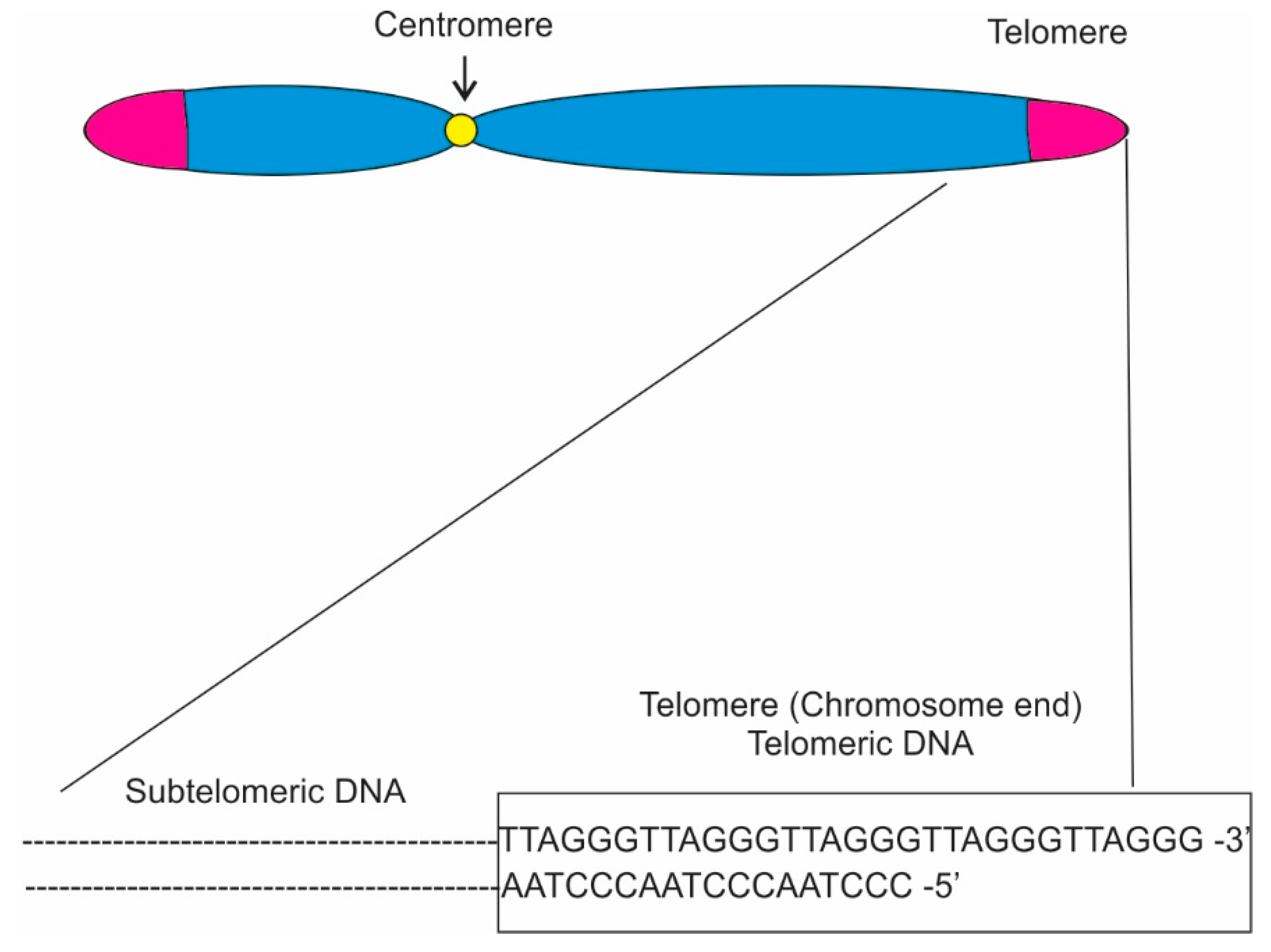
Lastly, the budding yeast genome contains one single rDNA cluster arranged as a linear array of 150–200 rDNA repeats on chromosome XII Fission yeast contain two rDNA arrays composed of 80–120 rDNA repeats located at the two ends of chromosome 3.
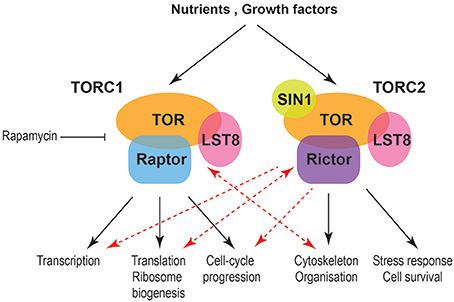





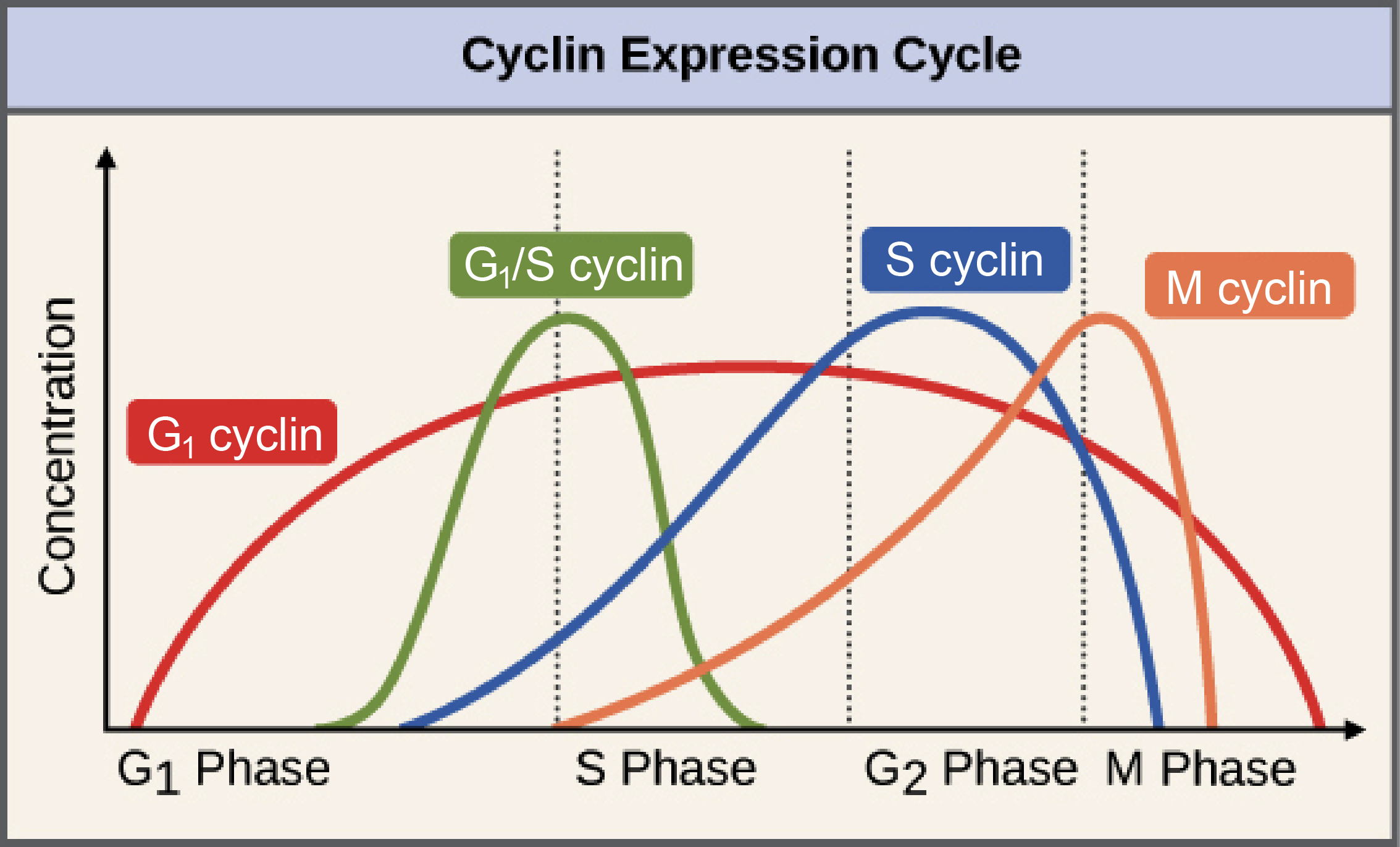
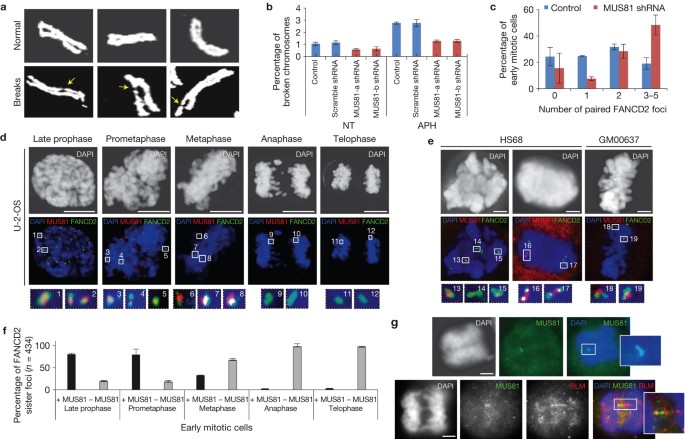
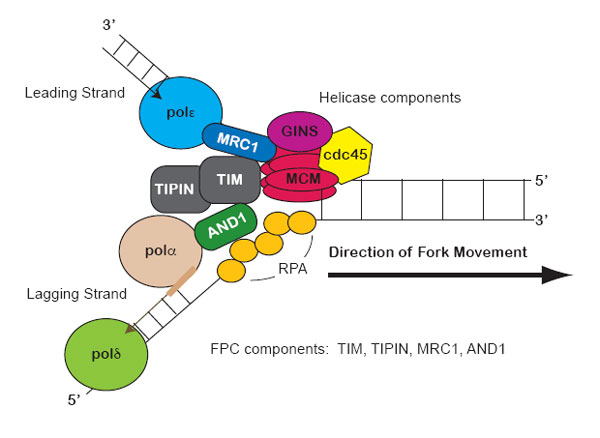 Stalled replication forks produce physical changes to DNA that induce checkpoint signals. For example, the minichromosome maintenance (MCM) helicase continues unwinding the DNA and generates some excess single stranded DNA (ssDNA).
Stalled replication forks produce physical changes to DNA that induce checkpoint signals. For example, the minichromosome maintenance (MCM) helicase continues unwinding the DNA and generates some excess single stranded DNA (ssDNA).
 Because the events of MiDAS were determined to occur after G2 but before prometaphase, the authors were interested to see what in prophase contributed to new DNA synthesis.
Because the events of MiDAS were determined to occur after G2 but before prometaphase, the authors were interested to see what in prophase contributed to new DNA synthesis. (A) thymidine amino acid, (B) 5-bromo-2'-deoxyuridine antibody (BrdU), and (C) 5-ethynyl-2'-deoxyuridine azide (EdU).
(A) thymidine amino acid, (B) 5-bromo-2'-deoxyuridine antibody (BrdU), and (C) 5-ethynyl-2'-deoxyuridine azide (EdU).