ToDo/@lma@mstdn.social
ToDoBioinformacion
@lma@mstdn.social
ToDoBioinformacion
@lma@mstdn.social
BIOmastodonte
ArticuloGenial
PCM
HerramientasPubMed
BIOrss
BIOia
AzulSemantico
AzulSemantico
RevisionBIOcolores
AzulSemantico
UnColor
VertebradosBIOColores
https://www.zotero.org/lmichan/collections/J6GEZUVI/items/SVJPNG2K/item-list
AzulSemantico
mlbb_biodiversidad
BIOnombres
QueEsElSexo
VertebradosBIOcolores
Kit_metria
Valdría la pena agregar Lens.org
Kit_revisiones
RevisionBIOcolores
EcologíaBIOcolores
ArañasBIOColores
https://www.zotero.org/lmichan/collections/J6GEZUVI/items/LQJQ8EEZ/item-list
Kit_metria
RevisionBIOcolores
OrqganosBIOcolores
NegroBIOcolores
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/7RU3CNZ4/item-list
GrafosConocimiento
Descripción: Aplicación Web abierta para visualizar relaciones de citación de los artículos científicos
ModeloBiocolores
ColibriesBIOColores
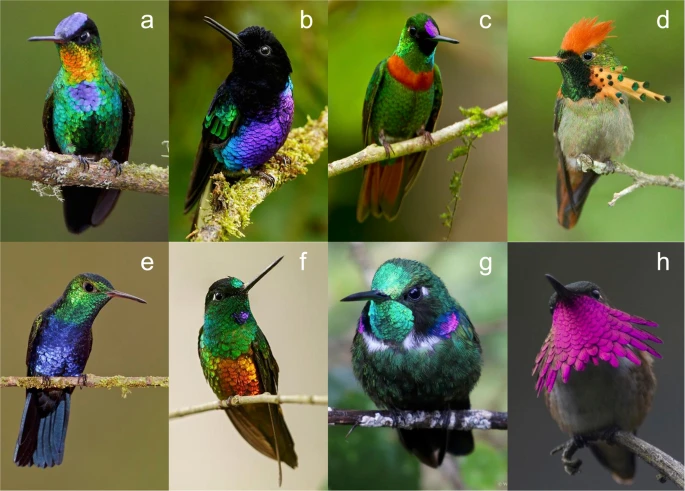
PaletasBIOcolores
BancoImagenes
MAriposasBIOcolores
PolimorfismoBIOColores
AlgodonBIOColores
RevisionBIOcolores
ReptilesBIOcolores
https://www.zotero.org/lmichan/collections/J6GEZUVI/items/WTBBPS3Y/item-list
Proyecto "Anotación PFR"
https://github.com/lmichan/PFR
TipoDePrueba/mesh/D002000/ForcedSpirometry
RevisionBIOcolores
AvesBIOcolores
LorosBIOcolores
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/G69YW698/item-list
RevisionBIOcolores
GeneticaBIOColores
AnimalesBIOcolores
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/GBXETNMC/item-list
ModeloBiocolores
BiocoloresSciBot
LorosBIOColores
AmarilloBIOcolores
Este es un bonito ejemplo de coloración resultado de pigmentación y color estructural en aves

https://docs.google.com/presentation/d/1JBWiw4_E06UW0uiwGurK7i52iSJZCIx4I--RLY7LNdY/edit?usp=sharing
BIOmuseos
RevisionBIOcolores
VariablesBIOColores
🚩U+1F6A9 FocusedReview
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/HVC3KZVP/item-list
BIOwikidata
BibliografíaWikidata
BibliografíaWikidata
BibliografíaWikidata
BibliografíaWikidata
BIOzotero
AvesBIOcolores
OjosBIOColores
CarotenoidesBIOcolores
RevisionBIOcolores
MariposasBIOcolores
EyspotsBIOColores
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/2XD9JFAB/item-list
RevisionBIOcolores
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/9QV5Y7KJ/item-list
RevisionBIOcolores
FrutosBIOcolores
FresasBIOColores
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/ENQBSAKT/item-list
RevisionBIOcolores
tipo de revisión
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/FZV5M5E8/item-list
RevisionBIOcolores
FloresBIOcolores
AzulElusivo
AzulSemántico
AzulBiocolores
UnColor
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/GV7VYCV9/item-list
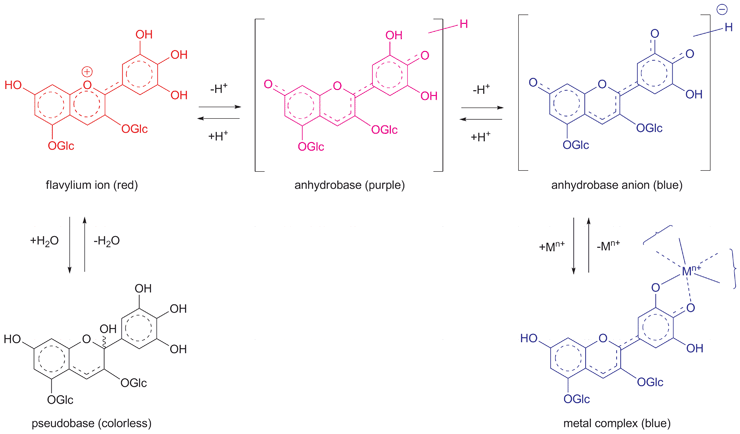
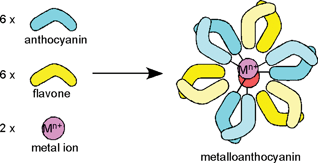
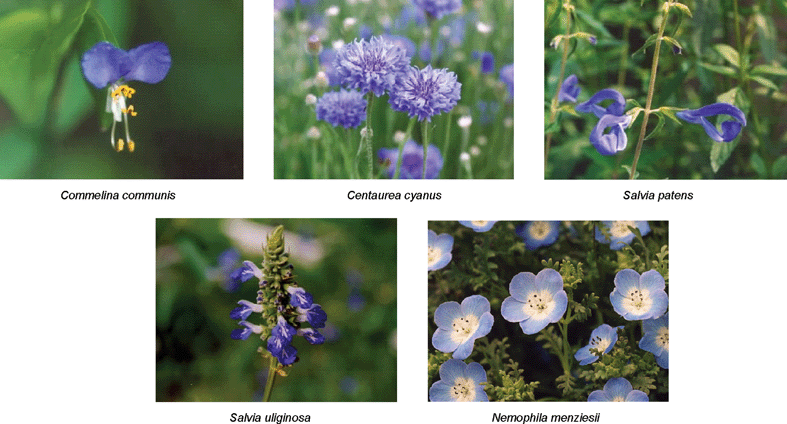
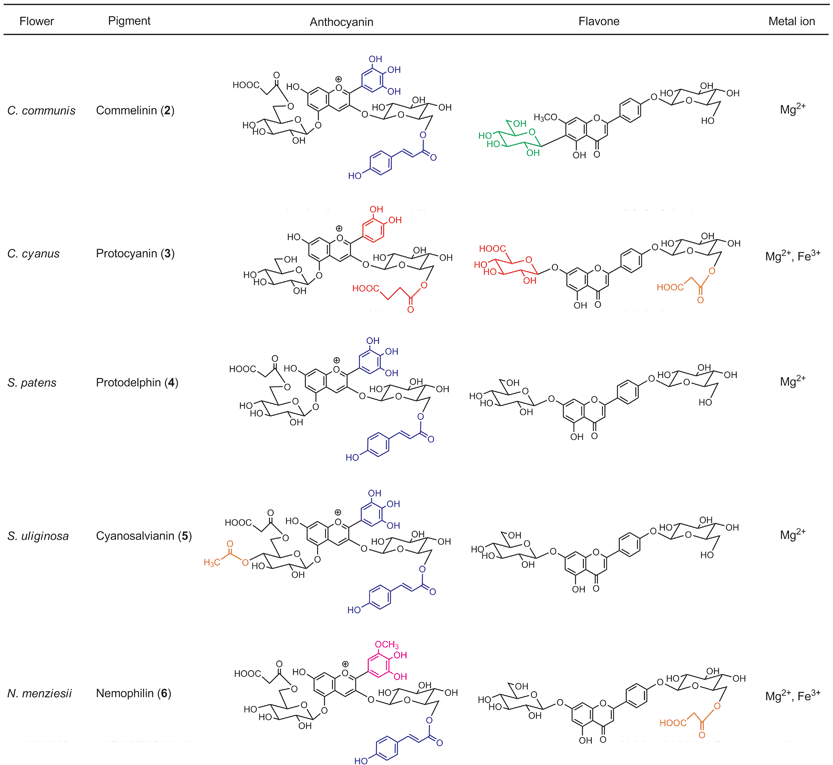
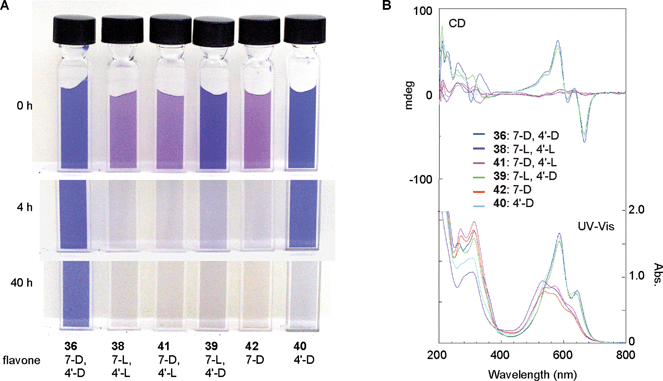
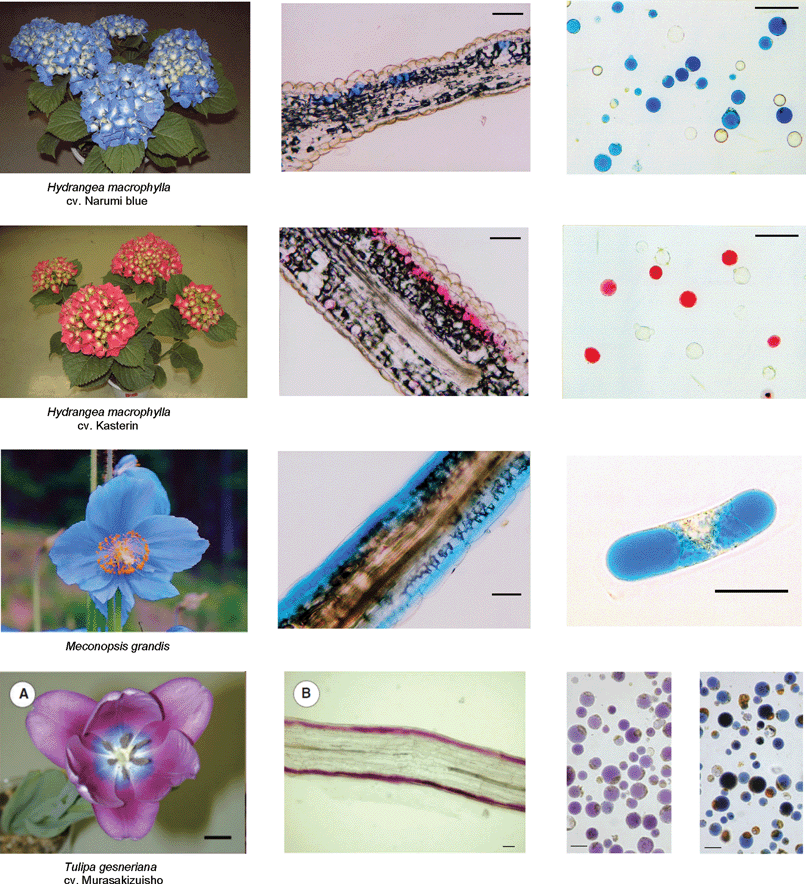
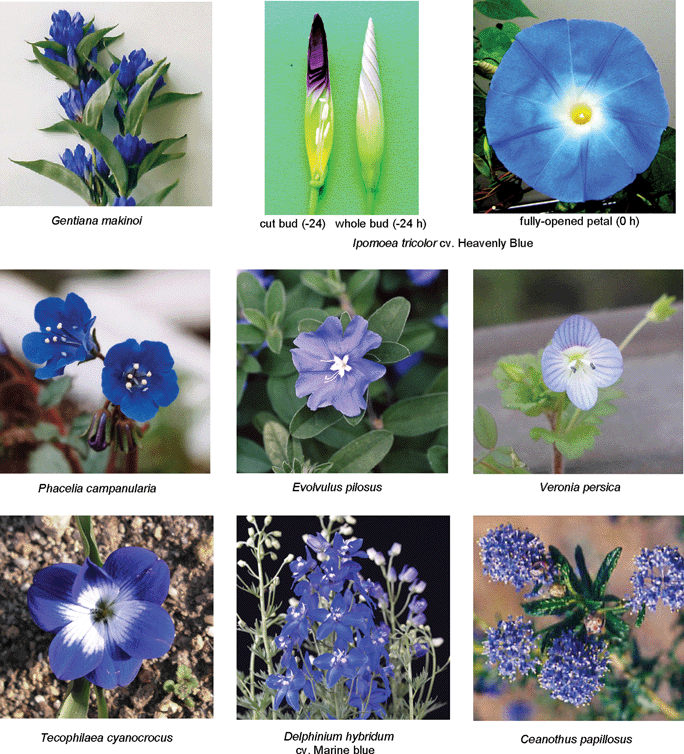
RevisionBIOcolores
PigmentosBIocolores
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/VGMH8CET/item-list
Las células pigmentarias son un buen modelo para estudiar el transporte celular
RevisionBIOColores
EcologíaBIOcolores
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/MVZ8W96S/item-list
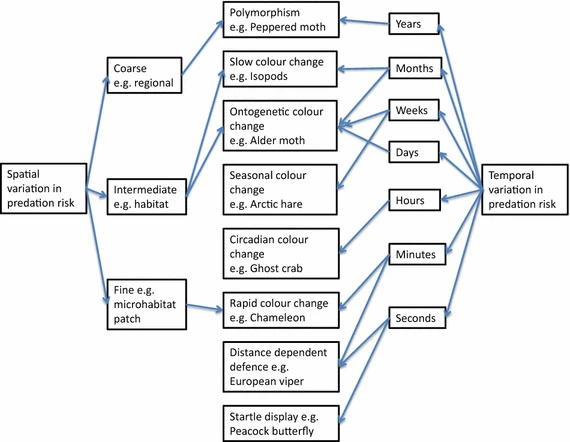
ColorEstructuralBiocolores
AzulBIocolores
IridiscenciaBiocolores
VariableBiocolores
RevisionBIOcolores
Marcar todas las plantas con iridiscencia y color estructural
act:todo
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/ZKXM854I/item-list


RevisionBIOcolores
AposematismoBIOColores
https://www.zotero.org/lmichan/collections/J6GEZUVI/items/C2MI3LCT/item-list
RevisionBIOcolores
BIOdiseñoBIOColores
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/X5WKF5AR/item-list
RevisionBIOcolores
BIOmetaanalisis
RevisionBIOcolores
EyespotsBIOColores
MariposasBIOColores
Es revision?
MariposasBIOColores
EyespotsBIOColores
RevisionBIOcolores
OjosBIOColores
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/AUPWMBAE/item-list
RevisionBIOcolores
MariposasBIOColores
EyespotsBIOColores
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/AAL7GYL5/item-list
RevisionBIOcolores
AnelidosBIOcolores
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/PWE374JN/item-details
RevisionBIOcolores
OjosBIOColores
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/FWX3WBST/item-details
RevisionBIOcolores
MariposasBIOColores
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/RUD6ZZM5/item-details
RevisionBIOcolores
OjosBIOcolores
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/4T432NQS/item-list
RevisionBIOcolores
Es revision?
RevisionBIOcolores
OjosBIOColores
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/2FNKCXW7/item-list
RevisionBIOcolores
MariposasBIOColores
Es revisión?
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/DBWEPUAG/item-list
RevisionBIOcolores
ModeloBiocolores
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/7FP74U36/item-details
RevisionBIOcolores
ModeloBiocolores
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/K8T8XVD8/item-details
Colage
DinosauriosBIOcolores
HuevosBIOColores
BIOrevistas
BIOrevistas
BIOdataviz
MamiferosBIOcolores
ArdillasBIOcolores
EvolucionBIOColores
JirafasBIOcolores
JirafasBIOcolores
HerramientasPubMed
HerramientasPubMed
HerramientasPubMed
HerramientasPubMed
HerramientasPubMed
HerramientasPubMed
HerramientasPubMed
HerramientasPubMed
HerramientasPubMed
HerramientasPubMed
HerramientasPubMed
HerramientasPubMed
HerramientasPubMed
HerramientasPubMed
HerramientasPubMed
https://github.com/lmichan/BioDBS
nar:https://academic.oup.com/nar/article/49/D1/D1/6059975
wd:https://www.wikidata.org/wiki/Q107654190
narid:(https://www.oxfordjournals.org/nar/database/summary/2261?action=Search§ion=all&term=litcovid
dc_subject: "COVID-19"
dc_date: 2021
HerramientaPubMed
HerramientaPubMed
bibliografia,ej/articulo_defrosting,cursoID/bibliografia
BIOliteratura
RevisionBIOcolores
https://www.zotero.org/groups/5400707/revisiones_biocolores/items/8NWQZ55K/item-details
metria
PCM
PCM20242
PCM
PCM20242
PCM
PCM20242
PCM
PCM20242
PCM
PCM20242
PCM
PCM20242
PCM
PCM20242
PCM
PCM20242
PCM
PCM20242
PCM
PCM20242
PCM
PCM20242
PCM
PCM20242
PCM
PCM20242
PCM
PCM20242
PCM
PCM20242
PCM
PCM20242
PCM
PCM20242
AlonsoGutiérrezRomero
PCM
PCM20242
Colage
Fluorescencia

PlantasBIOcolores
MimetismoBIOcolores
patrones de color floral
FormaFlor
VariablesBIOcolores

MariposasBIOcolores
TecnicaBIOcolores
MuseomicaBIOcolores



TecnicaBIOcolores
MuseomicaBIOcolores
AvesBIOcolores
https://twitter.com/i/status/1765393047653933378
ConosBIOcolores
BuenasPracticas
MariposasBIOcolores
Colage
CatalogoBIODBS
CatalogoBIODBS
CatalogoBIODBS
OrugasAzules
OrganismosAzules
CatalogoBIODBS
CatalogoBIODBS
CatalogoBIODBS
BIOmapasDeConocimiento
BIOgrafosConocimiento
CatalogoBIODBS
CatalogoBIODBS
CatalogoBIODBS
CatalogoBIODBS
DirectorioBIO-ontologias
BIOespecificaciones
CatalogoBIODBS
CatalogoBIODBS
FormaFlor
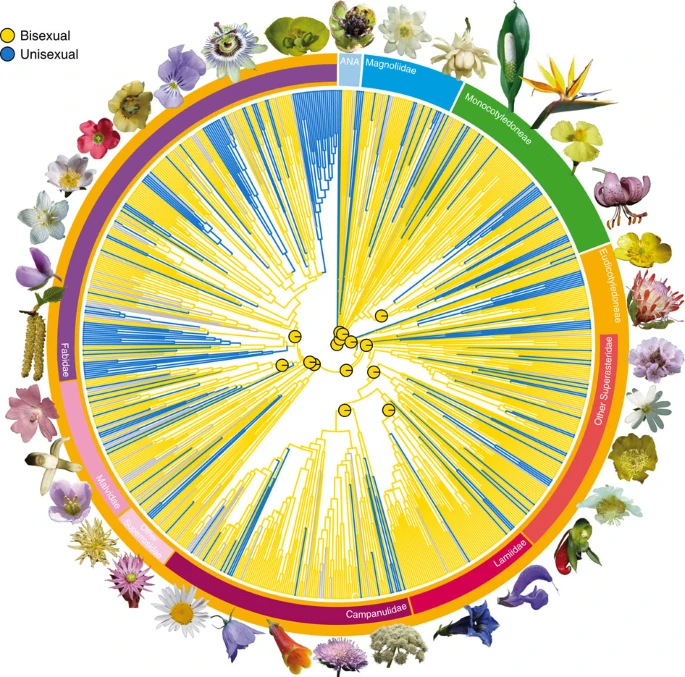
OjosBIOcolores
GeneticaBIOcolores
FenotipoBIOcolores
AvesBIOcolores
ProyectosBIOcolores
BIOgrafosConocimiento
En las últimas décadas, los biólogos evolutivos han mejorado enormemente los métodos computacionales para agilizar la recopilación y el análisis de datos genéticos de sistemas no modelo. Sin embargo, las herramientas para datos fenotípicos están rezagadas a pesar de la necesidad crítica de dichos datos para comprender cómo han evolucionado los rasgos a escalas micro y macroevolutivas. En comparación con los datos de las bases de datos genéticas estandarizadas (NCBI, etc.), los datos fenotípicos a menudo se encuentran dispersos en repositorios no estándar (por ejemplo, material complementario), no están disponibles públicamente o no son comparables entre estudios debido a la subjetividad de los términos utilizados. incapacidad para probar la homología dados los datos faltantes y variabilidad de los enfoques para codificar caracteres y estados de caracteres. La terminología subjetiva para caracterizar fenotipos obstaculiza nuestra capacidad de aprovechar los datos existentes para metanálisis a gran escala en investigaciones evolutivas o morfológicas o determinar si los resultados de los estudios son comparables. Estos problemas se magnifican en estudios que incluyen morfologías complejas o mal caracterizadas donde la terminología es a menudo imprecisa y conflictiva, o específica del grupo de estudio. Para aumentar la claridad de los datos y la reproducibilidad científica, abogamos por el uso de ontologías existentes y la mejora continua de las ontologías vegetales.
TO DO - Leer [ ]

