8,037 Matching Annotations
- Jan 2022
-
solid.github.io solid.github.io
-
github.com github.com
-
-
hal.archives-ouvertes.fr hal.archives-ouvertes.fr
-
Abstract : Current tag modelling does not fully take into account the rich and diverse nature tags, as signs, can take on. We propose an ontology of tags in which tags are modelled as named graphs. These named graphs are made of a resource linked to a “sign” which can be any resource reachable on the Web (an ontology concept, an image, etc.). The purpose of our model is to be able to describe tags in a very general manner, and as an immediate conse- quence, to describe tags as modelled by other tag models (SCOT, CommonTag, etc.).
Tags
Annotators
URL
-
-
ns.inria.fr ns.inria.fr
Tags
Annotators
URL
-
-
fr.slideshare.net fr.slideshare.net
-
webdesign.tutsplus.com webdesign.tutsplus.com
-
www.flickr.com www.flickr.com
-
rowanmanning.com rowanmanning.com
-
betterprogramming.pub betterprogramming.pub
-
-
-
threads.js.org threads.js.org
-
-
mmazzarolo.com mmazzarolo.com
-
A note on setting worker-loader’s publicPath with webpack 5
Webpack 5 introduced a mechanism to detect the publicPath that should be used automatically
[...]
Webpack 5 exposes a global variable called
__webpack_public_path__that allows you to do that.// Updates the `publicPath` at runtime, overriding whatever was set in the // webpack's `output` section. __webpack_public_path__ = "/workers/"; const myWorker = new Worker( new URL("/workers/worker.js"); ); // Eventually, restore the `publicPath` to whatever was set in output. __webpack_public_path__ = "https://my-static-cdn/";
-
-
-
{ "@context": { "doap": "http://usefulinc.com/ns/doap#", "url": "@id", "name": "doap:name", "description": "doap:description", "author": "doap:maintainter", "license": "doap:license", // can we map values to https://spdx.org/licenses/ ? ... }, "homepage": {"@id": "doap:homepage", "@type": "@id"} }
-
-
Tags
Annotators
URL
-
-
dzone.com dzone.com
Tags
Annotators
URL
-
-
rustwasm.github.io rustwasm.github.io
-
www.w3.org www.w3.org
-
This document contains information about embedding metadata in W3C Technical Reports (TR) using RDFa.
-
-
hypothes.is hypothes.is
-
-
hypothes.is hypothes.is
-
-
-
-
example.com example.com
-
Here’s an even more magical trick. Download that PDF to your file system, load it into a third tab, and annotate again. Now you’ll see all three annotations in all three tabs!
Since Hypothesis doesn’t know that the local copy of the PDF came from http://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0168597&type=printable, or that it’s related to http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0168597, how is that possible?
The answer is that the PDF standard defines a unique identifier, or “fingerprint,” that authoring tools encode into the PDFs they create. When you use the Hypothesis client to annotate web-hosted PDF, it captures the fingerprint and sends it to the server.
-
It was already the case that you could search Hypothesis for the DOI, like so:
-
First, here’s a magic trick you might not realize Hypothesis has up its sleeve. Consider this PLOS One article. Annotate it in one tab, then open a second tab and annotate the PDF version there. You’ll see both annotations in both tabs. How is that possible?
The answer is that when scholarly publishers provide HTML versions of articles, they typically include metadata that points to PDF versions of the same articles. Here’s one way that happens:
<meta name=”citation_pdf_url” content=”http://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0168597&type=printable”>Hypothesis remembers the correspondence between the HTML and PDF versions, and coalesces annotations across them.
-
Tags
Annotators
URL
-
-
vocabularies.coar-repositories.org vocabularies.coar-repositories.org
Tags
Annotators
URL
-
-
-
This specification defines two formats and respective media types for representing sets of links as stand-alone documents. One format is JSON-based, the other aligned with the format for representing links in the HTTP "Link" header field. This specification also introduces a link relation type to support discovery of sets of links.
GET links/resource1 HTTP/1.1 Host: example.org Accept: application/linkset+jsonHTTP/1.1 200 OK Date: Mon, 12 Aug 2019 10:46:22 GMT Server: Apache-Coyote/1.1 Content-Type: application/linkset+json Link: <https://example.org/links/resource1> ; rel="alternate" ; type="application/linkset" Content-Length: 1349 { "linkset": [ { "anchor": "https://example.org/resource1", "author": [ { "href": "https://authors.example.net/johndoe", "type": "application/rdf+xml" } ], "memento": [ { "href": "https://example.org/resource1?version=1", "type": "text/html", "datetime": "Thu, 13 Jun 2019 09:34:33 GMT" }, { "href": "https://example.org/resource1?version=2", "type": "text/html", "datetime": "Sun, 21 Jul 2019 12:22:04 GMT" } ], "latest-version": [ { "href": "https://example.org/resource1?version=3", "type": "text/html" } ] }, { "anchor": "https://example.org/resource1?version=3", "predecessor-version": [ { "href": "https://example.org/resource1?version=2", "type": "text/html" } ] }, { "anchor": "https://example.org/resource1?version=2", "predecessor-version": [ { "href": "https://example.org/resource1?version=1", "type": "text/html" } ] }, { "anchor": "https://example.org/resource1#comment=1", "author": [ { "href": "https://authors.example.net/alice"} ] } ] }
Tags
Annotators
URL
-
-
robustlinks.mementoweb.org robustlinks.mementoweb.org
Tags
Annotators
URL
-
-
robustlinks.mementoweb.org robustlinks.mementoweb.org
-
4. Robustifying a link when linking to a specific version
If the main intent is to link to a specific state of an original resource, for example a snapshot of the original resource in a web archive or one of its version in a version control system, then Robust Link information is conveyed as follows:
hreffor the URI that provides the specific state i.e., the snapshot or resource version;data-originalurlfor the URI of the original resource;data-versiondatefor the datetime of the snapshot or resource version.
[...]
<a href="http://en.wikipedia.org/w/index.php?title=Web_archiving&oldid=485347845" data-originalurl="http://en.wikipedia.org/wiki/Web_archiving" data-versiondate="2012-03-20">Robust Link to this specific version of the Wikipedia page</a> -
3. Robustifying a link when linking to the original resource
If the main intent is to link to an original resource but also allow future users of that link to see the state of the original resource around the time the link was put in place, then Robust Link information is conveyed as follows:
hreffor the URI of the original resource for which the snapshot was taken;data-versionurlfor the URI of the snapshot;data-versiondatefor the datetime of linking, of taking the snapshot.
[...]
<a href="http://www.w3.org/" data-versionurl="https://archive.today/r7cov" data-versiondate="2015-01-21">Robust Link to the W3C home page</a> -
The approach proposed here is to convey this information on a link by leveraging HTML5's attribute extensibility mechanism. It introduces the following
data-attributes for the anchor (<a>) element:data-originalurlfor the URI of the original resource;data-versionurlfor the URI of the snapshot;data-versiondatefor the datetime of linking, of taking the snapshot.
-
Robust Links provide multiple pathways to revisit a link's original content, even a long time after the link was put in place. This document describes approaches to robustify links in HTML pages. All approaches assume that, when linking to a web resource, a snapshot of the state of that resource is created, for example, in a web archive or a versioning system. When linking, the URI of the resource, the URI of the snapshot, and the datetime of linking are conveyed.
Tags
Annotators
URL
-
-
hypothes.is hypothes.is
-
-
twitter.com twitter.comTwitter1
-
twitter.com twitter.com
-
docs.genius.com docs.genius.com
-
-
css-tricks.com css-tricks.com
Tags
Annotators
URL
-
-
pomax.github.io pomax.github.io
Tags
Annotators
URL
-
-
tools.ietf.org tools.ietf.org
Tags
Annotators
URL
-
-
developer.twitter.com developer.twitter.com
-
www.whosampled.com www.whosampled.com
-
thepund.it thepund.it
-
-
-
-
stackoverflow.com stackoverflow.com
-
String.split()can also accept a regular expression:input.split(/[ ,]+/);
-
-
datatracker.ietf.org datatracker.ietf.org
-
This document describes a Uniform Resource Name (URN) Namespace Identification (NID) convention as prescribed by the Internet Engineering Task Force (IETF) for identifying, naming, assigning, and managing persistent resources in the legal domain.
Tags
Annotators
URL
-
-
developers.google.com developers.google.com
-
copilot.github.com copilot.github.com
Tags
Annotators
URL
-
-
sethmlarson.dev sethmlarson.dev
Tags
Annotators
URL
-
-
levelup.gitconnected.com levelup.gitconnected.com
-
stackoverflow.com stackoverflow.com
-
hypothes.is hypothes.is
-
-
-
-
thoughtspile.github.io thoughtspile.github.io
Tags
Annotators
URL
-
-
-
-
stackoverflow.com stackoverflow.com
-
b) rgb(), rgba(), hsl() and hsla():
^(rgb|hsl)(a?)[(]\s*([\d.]+\s*%?)\s*,\s*([\d.]+\s*%?)\s*,\s*([\d.]+\s*%?)\s*(?:,\s*([\d.]+)\s*)?[)]$
-
-
www.reddit.com www.reddit.com
-
hypothes.is hypothes.is
-
-
-
<link rel="prefetch" href="/style.css" as="style" /> <link rel="preload" href="/style.css" as="style" /> <link rel="preconnect" href="https://example.com" /> <link rel="dns-prefetch" href="https://example.com" /> <link rel="prerender" href="https://example.com/about.html" /> <link rel="modulepreload" href="/script.js" />
-
-
www.executeprogram.com www.executeprogram.com
Tags
Annotators
URL
-
-
stackoverflow.com stackoverflow.com
-
remix.guide remix.guide
-
-
blog.saeloun.com blog.saeloun.com
-
developer.chrome.com developer.chrome.com
Tags
Annotators
URL
-
-
hypothes.is hypothes.is
-
-
ibotpeaches.github.io ibotpeaches.github.io
-
-
tinybase.org tinybase.orgTinyBase1
-
-
jbiomedsem.biomedcentral.com jbiomedsem.biomedcentral.com
-
stackoverflow.com stackoverflow.com
-
epress.trincoll.edu epress.trincoll.edu
Tags
Annotators
URL
-
-
ncsu-libraries.github.io ncsu-libraries.github.io
Tags
Annotators
URL
-
-
webconcepts.info webconcepts.info
-
-
cloud.google.com cloud.google.com
-
slides.com slides.com
-
docs.google.com docs.google.com
-
clariah.github.io clariah.github.io
-
-
-
mermaid-js.github.io mermaid-js.github.io
-
Mermaid lets you create diagrams and visualizations using text and code.It is a Javascript based diagramming and charting tool that renders Markdown-inspired text definitions to create and modify diagrams dynamically.
Tags
Annotators
URL
-
-
github.com github.com
-
```mermaid graph TD; A-->B; A-->C; B-->D; C-->D;```

Tags
Annotators
URL
-
-
www.openarchives.org www.openarchives.org
Tags
Annotators
URL
-
-
pro.europeana.eu pro.europeana.eu
-
The Annotations API is an extension to the Europeana REST API which allows you to create, retrieve and manage annotations on Europeana objects. Annotations are user-contributed or system-generated enhancements, additions or corrections to (or a selection of) metadata or media. We adopted the Web Annotation Data Model as a base model for the representation of annotations and as a format for exchanging annotations between client applications and the API, but also the Web Annotation Protocol as base HTTP protocol for the API.
Example:
{ "@context": “http://www.w3.org/ns/anno.jsonld” "id": "http://data.europeana.eu/annotations/1", "type": "Annotation", "created": "2015-03-10T14:08:07Z", "creator": { "type": "Person", "name": "John Smith" }, "generated": "2015-04-01T09:00:00Z", "generator": { "type": "Software", "name": "HistoryPin", "homepage": "https://www.historypin.org/" }, "motivation": "tagging", "bodyValue": "MyBeautifulTag", "target": "http://data.europeana.eu/item/92062/BibliographicResource_1000126189360" }
Tags
Annotators
URL
-
-
linked.art linked.artAPI1
Tags
Annotators
URL
-
-
iphylo.blogspot.com iphylo.blogspot.com
-
This is the profile for Mark Hughes (0000-0002-2168-0514). Up until now I've been generating my own linked data version of ORCID records that look very similar to this, but going forward this will simplify life.
Note that I've truncated the above example, it's actually this:
{ "@context": "http://schema.org", "@type": "Person", "@id": "https://orcid.org/0000-0002-2168-0514", "mainEntityOfPage": "https://orcid.org/0000-0002-2168-0514", "givenName": "Mark", "familyName": "Hughes", "affiliation": { "@type": "Organization", "name": "Royal Botanic Garden Edinburgh", "identifier": { "@type": "PropertyValue", "propertyID": "RINGGOLD", "value": "41803" } }, "@reverse": { "creator": [ { "@type": "CreativeWork", "@id": "https://doi.org/10.1017/s0960428619000283", "name": "BEGONIA MAGUNIANA (BEGONIACEAE, BEGONIA SECT. OLIGANDRAE), A NEW SPECIES FROM NEW GUINEA", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1017/s0960428619000283" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.11646/phytotaxa.407.1.11", "name": "A revision of Begonia sect. Petermannia on Sumatra, Indonesia", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.11646/phytotaxa.407.1.11" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.11646/phytotaxa.407.1.4", "name": "Two new species of Begonia (Begoniaceae) from Borneo", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.11646/phytotaxa.407.1.4" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1017/s0960428619000052", "name": "AN UPDATED CHECKLIST AND A NEW SPECIES OF BEGONIA (B. RHEOPHYTICA) FROM MYANMAR", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1017/s0960428619000052" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1371/journal.pone.0194877", "name": "Chloroplast and nuclear DNA exchanges among Begonia sect. Baryandra species (Begoniaceae) from Palawan Island, Philippines, and descriptions of five new species.", "identifier": [ { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1371/journal.pone.0194877" }, { "@type": "PropertyValue", "propertyID": "pmc", "value": "PMC5931476" }, { "@type": "PropertyValue", "propertyID": "pmid", "value": "29718922" } ], "sameAs": [ "https://europepmc.org/articles/PMC5931476", "https://www.ncbi.nlm.nih.gov/pubmed/29718922" ] }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1017/s0960428618000136", "name": "TWO NEW SPECIES OF BEGONIA FROM SUMATRA", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1017/s0960428618000136" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1017/s096042861800001x", "name": "A REVISION OF BEGONIA SECT. SYMBEGONIA ON NEW GUINEA", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1017/s096042861800001x" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.5852/ejt.2018.396", "name": "A revision and one new species of Begonia L. (Begoniaceae, Cucurbitales) in Northeast India", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.5852/ejt.2018.396" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1002/tax.606013", "name": "Pliocene intercontinental dispersal from Africa to Southeast Asia highlighted by the new species Begonia afromigrata (Begoniaceae)", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1002/tax.606013" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.11646/phytotaxa.381.1.16", "name": "Taxonomic notes on the Philippine endemic Begonia colorata (Begoniaceae, section Petermannia)", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.11646/phytotaxa.381.1.16" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1186/s40529-017-0182-x", "name": "Three new species of Begonia sect. Baryandra from Panay Island, Philippines.", "identifier": [ { "@type": "PropertyValue", "propertyID": "pmid", "value": "28664395" }, { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1186/s40529-017-0182-x" }, { "@type": "PropertyValue", "propertyID": "pmc", "value": "PMC5491425" } ], "sameAs": [ "https://www.ncbi.nlm.nih.gov/pubmed/28664395", "https://europepmc.org/articles/PMC5491425" ] }, { "@type": "CreativeWork", "@id": "https://doi.org/10.3767/000651917x695083", "name": "A new species of Begonia section Parvibegonia (Begoniaceae) from Thailand and Myanmar", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.3767/000651917x695083" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1017/s0960428617000075", "name": "TAXONOMY OF BEGONIA ALBOMACULATA AND DESCRIPTION OF TWO NEW SPECIES ENDEMIC TO PERU", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1017/s0960428617000075" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1017/s0960428617000051", "name": "FOUR NEW SPECIES OF BEGONIA (BEGONIACEAE) FROM THAILAND", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1017/s0960428617000051" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.3850/s2382581216000077", "name": "A new species and a new record in Begonia sect. Platycentrum (Begoniaceae) from Thailand", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.3850/s2382581216000077" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.5852/ejt.2015.119", "name": "Begonia yapenensis (sect. Symbegonia, Begoniaceae), a new species from Papua, Indonesia", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.5852/ejt.2015.119" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.11646/phytotaxa.197.1.4", "name": "A new section (Begonia sect. Oligandrae sect. nov.) and a new species (Begonia pentandra sp. nov.) in Begoniaceae from New Guinea", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.11646/phytotaxa.197.1.4" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.5852/ejt.2015.167", "name": "Further discoveries in the ever-expanding genus Begonia (Begoniaceae): fifteen new species from Sumatra", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.5852/ejt.2015.167" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1186/s40529-015-0099-1", "name": "Three New Species of Begonia Endemic to the Puerto Princesa Subterranean River National Park, Palawan", "identifier": [ { "@type": "PropertyValue", "propertyID": "other-id", "value": "al:1817406x-201507-201507290029-201507290029-c1-14" }, { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1186/s40529-015-0099-1" } ], "sameAs": "http://www.airitilibrary.com/Publication/alDetailedMesh?DocID=1817406X-201507-201507290029-201507290029-c1-14" }, { "@type": "CreativeWork", "@id": "https://doi.org/10.5852/ejt.2013.56", "name": "Memecylon pseudomegacarpum M.Hughes (Melastomataceae), a new species of tree from Peninsular Malaysia", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.5852/ejt.2013.56" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1186/1999-3110-54-38", "name": "Recircumscription of Begonia sect. Baryandra (Begoniaceae): evidence from molecular data", "identifier": [ { "@type": "PropertyValue", "propertyID": "other-id", "value": "al:1817406x-201309-201401170003-201401170003-70-74" }, { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1186/1999-3110-54-38" } ], "sameAs": "http://www.airitilibrary.com/Publication/alDetailedMesh?DocID=1817406X-201309-201401170003-201401170003-70-74" }, { "@type": "CreativeWork", "@id": "https://doi.org/10.11646/phytotaxa.66.1.2", "name": "A new species and new combinations of Memecylon in Thailand and Peninsular Malaysia", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.11646/phytotaxa.66.1.2" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1017/s0960428612000078", "name": "A NEW SPECIES OF BEGONIA (BEGONIACEAE) FROM PENINSULAR THAILAND", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1017/s0960428612000078" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.3329/bjpt.v19i2.13134", "name": "Pollen morphology of Begonia L. (Begoniaceae) in Nepal", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.3329/bjpt.v19i2.13134" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1111/j.1365-2699.2011.02596.x", "name": "West to east dispersal and subsequent rapid diversification of the mega-diverse genus Begonia (Begoniaceae) in the Malesian archipelago", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1111/j.1365-2699.2011.02596.x" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1017/s0960428611000072", "name": "NINE NEW SPECIES OF BEGONIA (BEGONIACEAE) FROM SOUTH AND WEST SULAWESI, INDONESIA", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1017/s0960428611000072" } }, { "@type": "CreativeWork", "name": "Begonia blancii (sect. Diploclinium, Begoniaceae), A New Species Endemic to the Philippine Island of Palawan", "identifier": { "@type": "PropertyValue", "propertyID": "other-id", "value": "al:1817406x-201104-201106150032-201106150032-203-209" }, "sameAs": "http://www.airitilibrary.com/Publication/alDetailedMesh?DocID=1817406X-201104-201106150032-201106150032-203-209" }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1017/s0960428609005307", "name": "BEGONIA SECTION PETERMANNIA (BEGONIACEAE) ON PALAWAN (PHILIPPINES), INCLUDING TWO NEW SPECIES", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1017/s0960428609005307" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1017/s0960428609005484", "name": "TWO NEW SPECIES OF BEGONIA (BEGONIACEAE) FROM SOUTH SULAWESI, INDONESIA", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1017/s0960428609005484" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1017/s0960428609005320", "name": "TWO NEW SPECIES OF BEGONIA (BEGONIACEAE) FROM CENTRAL SULAWESI, INDONESIA", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1017/s0960428609005320" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1017/s096042860800509x", "name": "BEGONIA VARIPELTATA(BEGONIACEAE): A NEW PELTATE SPECIES FROM SULAWESI, INDONESIA", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1017/s096042860800509x" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1017/s0960428607000777", "name": "BEGONIA CLADOTRICHA (BEGONIACEAE): A NEW SPECIES FROM LAOS", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1017/s0960428607000777" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1017/s0960428606000588", "name": "FOUR NEW SPECIES OF BEGONIA (BEGONIACEAE) FROM SULAWESI", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1017/s0960428606000588" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1600/0363644054782297", "name": "A Phylogeny of Begonia Using Nuclear Ribosomal Sequence Data and Morphological Characters", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1600/0363644054782297" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1016/s0006-3207(02)00375-0", "name": "Population genetic structure in the endemic Begonia of the Socotra archipelago", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1016/s0006-3207(02)00375-0" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1017/s0960428602000082", "name": "A NEW ENDEMIC SPECIES OF BEGONIA (BEGONIACEAE) FROM THE SOCOTRA ARCHIPELAGO", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1017/s0960428602000082" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1046/j.1471-8286.2002.00201.x", "name": "Isolation of polymorphic microsatellite markers for Begonia sutherlandii Hook. f.", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1046/j.1471-8286.2002.00201.x" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.1046/j.1471-8286.2002.00185.x", "name": "Polymorphic microsatellite markers for the Socotran endemic herb Begonia socotrana", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.1046/j.1471-8286.2002.00185.x" } }, { "@type": "CreativeWork", "@id": "https://doi.org/10.3126/botor.v7i0.4386", "name": "Distribution Patterns of Begonia species in the Nepal Himalaya", "identifier": { "@type": "PropertyValue", "propertyID": "doi", "value": "10.3126/botor.v7i0.4386" } } ] }, "url": [ "https://www.rbge.org.uk/about-us/organisational-structure/staff/tropical-diversity/dr-mark-hughes/", "https://www.mendeley.com/profiles/mark-hughes/" ], "identifier": [ { "@type": "PropertyValue", "propertyID": "Loop profile", "value": "845425" }, { "@type": "PropertyValue", "propertyID": "Loop profile", "value": "826408" } ] }This gives us a list of Mark's publications from ORCID. If there aren't any publications listed, the
@reverseproperty is empty. Note that@reverseis a JSON-LD trick that enables the JSON-LD document to include not only things linked from Mark's ORCID id (e.g., his name and affiliation) but also things his ORCID id is linked to (e.g., that he is the author of works such as https://doi.org/10.1017/s0960428619000283). -
You can get linked data in JSON-LD using content negotiation. If we send a request to https://orcid.org/0000-0002-2168-0514 with
"Accept: application/ld+json"we get back something like this:{ "@context": "http://schema.org", "@type": "Person", "@id": "https://orcid.org/0000-0002-2168-0514", "mainEntityOfPage": "https://orcid.org/0000-0002-2168-0514", "givenName": "Mark", "familyName": "Hughes", "affiliation": { "@type": "Organization", "name": "Royal Botanic Garden Edinburgh", "identifier": { "@type": "PropertyValue", "propertyID": "RINGGOLD", "value": "41803" } }, "@reverse": {}, "url": [ "https://www.rbge.org.uk/about-us/organisational-structure/staff/tropical-diversity/dr-mark-hughes/", "https://www.mendeley.com/profiles/mark-hughes/" ], "identifier": [ { "@type": "PropertyValue", "propertyID": "Loop profile", "value": "845425" }, { "@type": "PropertyValue", "propertyID": "Loop profile", "value": "826408" } ] }
-
-
developer.mozilla.org developer.mozilla.org
-
Tags
Annotators
URL
-
-
dublincore.org dublincore.org
-
-
github.com github.com
-
-
domiii.github.io domiii.github.io
-
-
maxleiter.com maxleiter.com
-
[...] In short, pinning dependencies means the exact version specified will be installed, rather than a dependency matching the range criteria. Here's an example:
{ "dependencies": { "react": "17.0.2", // installs react@17.0.2 exactly. I recommend this. "react": "^17.0.2", // installs the latest minor version after .0 (so 17.*.*) "react": "~17.0.2" // installs the latest patch after .2 (so 17.0.*) } }To automatically accomplish this in your projects, you can add
save-exact=trueto a.npmrcfile, or use--save-exactwhen adding the dependency via npm (or--exactvia yarn).
-
-
varun.ca varun.ca
-
-
-
Metadata for comic books and comic book collections.
-
-
www.joshwcomeau.com www.joshwcomeau.com
Tags
Annotators
URL
-
-
discovery.biothings.io discovery.biothings.io
Tags
Annotators
URL
-
-
hypothes.is hypothes.is
Tags
Annotators
URL
-
-
hypothes.is hypothes.is
-
-
blog.jonudell.net blog.jonudell.net
-
www.monperrus.net www.monperrus.net
-
scholar.google.com scholar.google.com
Tags
Annotators
URL
-
-
bibliontology.com bibliontology.com
-
-
apievangelist.com apievangelist.com
-
Here is what a base WebAPI type schema could look like:
{ "@context": "http://schema.org", "@type": "WebAPI", "name": "Google Knowledge Graph Search API", "description": "The Knowledge Graph Search API lets you find entities in the Google Knowledge Graph. The API uses standard schema.org types and is compliant with the JSON-LD specification.", "documentation": "https://developers.google.com/knowledge-graph/", "termsOfService": "https://developers.google.com/knowledge-graph/terms" }
-
-
openactive.io openactive.io
-
The below illustrates a Dataset Site pointing to feeds consisting of
ScheduledSessions,SessionSeries, andEvents. As the presence of thewebAPIattribute indicates, data items from these feeds are bookable.<script type="application/ld+json`/"> { "@context":[ "https://schema.org/", "https://openactive.io/", "https://openactive.io/ns-beta" ], "@type":"Dataset", "@id":"https://data.example.com/", "name":"Example Sessions and Events", "description":"Near real-time availability and rich descriptions relating to sessions and events available from Example.com", "url":"https://data.example.com/", "dateModified":"2019-08-25T11:23:27+00:00", "keywords":[ "Courses", "Sessions", "Events", "Activities", "Sports", "Physical Activity", "OpenActive" ], "schemaVersion":"https://www.openactive.io/modelling-opportunity-data/2.0/", "license":"https://creativecommons.org/licenses/by/4.0/", "publisher":{ "@type":"Organization", "name":"Example.com", "description":"Example.com makes it easy to get active!", "url":"https://example.com/home", "legalName":"Example Ltd", "logo":{ "@type":"ImageObject", "url":"https://cdn.example.com/assets/logo.png" }, "email":"support@example.com" }, "discussionUrl":"https://github.com/example/repo/issues", "datePublished":"2019-07-11T00:00:00+00:00", "inLanguage":[ "en-GB" ], "distribution":[ { "@type":"DataDownload", "name":"ScheduledSession", "additionalType":"https://openactive.io/ScheduledSession", "encodingFormat":"application/vnd.openactive.rpde+json; version=1", "contentUrl":"https://example.com/api/openactive/scheduledsessions", "totalItems": 1852 }, { "@type":"DataDownload", "name":"SessionSeries", "additionalType":"https://openactive.io/SessionSeries", "encodingFormat":"application/vnd.openactive.rpde+json; version=1", "contentUrl":"https://example.com/api/openactive/sessionseries", "totalItems": 361 }, { "@type":"DataDownload", "name":"Event", "additionalType":"https://schema.org/Event", "encodingFormat":"application/vnd.openactive.rpde+json; version=1", "contentUrl":"https://example.com/api/openactive/events", "totalItems": 1906 } ], "backgroundImage":{ "@type":"ImageObject", "url":"https://cdn.example.com/images/background.jpg" }, "documentation":"https://developer.openactive.io/", "accessService":{ "@type":"WebAPI", "name":"Open Booking API", "description":"The Open Booking API lets you to book OpenActive Opportunities. The API uses standard schema.org types and is compliant with the JSON-LD specification.", "documentation":"https://openactive.io/open-booking-api/EditorsDraft", "termsOfService":"https://example.com/api/booking/documentation/terms-of-service", "provider": { "@type": "Organization", "name":"examplebooking.com", "description":"examplebooking.com makes it easy to get booking!", "url":"https://examplebooking.com/home", "email":"support@examplebooking.com" }, "endpointUrl":"https://example.com/api/booking/", "conformsTo":[ "https://www.openactive.io/open-booking-api/2.0/" ], "endpointDescription":"https://www.openactive.io/open-booking-api/2.0/swagger.json", "bookingService": { "@type": "SoftwareApplication", "name": "nyExampleBookingPlatform", "softwareVersion": "1.2", "url": "https://www.example.com/myExampleBookingPlatform", "featureList": "https://www.example.com" } } } </script>
Tags
Annotators
URL
-
-
www.programmableweb.com www.programmableweb.com
Tags
Annotators
URL
-
-
github.com github.com
-
const pubpeer = await Zotero.HTTP.request('POST', 'https://pubpeer.com/v3/publications?devkey=PubPeerZotero', { body: JSON.stringify({ dois: fetch }), responseType: 'json', headers: { 'Content-Type': 'application/json;charset=UTF-8' }, })
-
-
addons.mozilla.org addons.mozilla.org
Tags
Annotators
URL
-
-
joshgoestoflatiron.medium.com joshgoestoflatiron.medium.com
-
function parseLinkHeader( linkHeader ) { const linkHeadersArray = linkHeader.split( ", " ).map( header => header.split( "; " ) ); const linkHeadersMap = linkHeadersArray.map( header => { const thisHeaderRel = header[1].replace( /"/g, "" ).replace( "rel=", "" ); const thisHeaderUrl = header[0].slice( 1, -1 ); return [ thisHeaderRel, thisHeaderUrl ] } ); return Object.fromEntries( linkHeadersMap ); }
-
-
medium.com medium.com
-
-
developer.mozilla.org developer.mozilla.org
-
tkdodo.eu tkdodo.eu
-
Data Transformation
[…]
3. using the select option
v3 introduced built-in selectors, which can also be used to transform data:
/* select-transformation */ export const useTodosQuery = () => useQuery(['todos'], fetchTodos, { select: (data) => data.map((todo) => todo.name.toUpperCase()), })selectors will only be called if data exists, so you don't have to care about undefined here. Selectors like the one above will also run on every render, because the functional identity changes (it's an inline function). If your transformation is expensive, you can memoize it either with
useCallback, or by extracting it to a stable function reference:/* select-memoizations */ const transformTodoNames = (data: Todos) => data.map((todo) => todo.name.toUpperCase()) export const useTodosQuery = () => useQuery(['todos'], fetchTodos, { // ✅ uses a stable function reference select: transformTodoNames, }) export const useTodosQuery = () => useQuery(['todos'], fetchTodos, { // ✅ memoizes with useCallback select: React.useCallback( (data: Todos) => data.map((todo) => todo.name.toUpperCase()), [] ), })Further, the select option can also be used to subscribe to only parts of the data. This is what makes this approach truly unique. Consider the following example:
/* select-partial-subscriptions */ export const useTodosQuery = (select) => useQuery(['todos'], fetchTodos, { select }) export const useTodosCount = () => useTodosQuery((data) => data.length) export const useTodo = (id) => useTodosQuery((data) => data.find((todo) => todo.id === id))Here, we've created a
useSelectorlike API by passing a custom selector to ouruseTodosQuery. The custom hooks still works like before, as select will be undefined if you don't pass it, so the whole state will be returned.But if you pass a selector, you are now only subscribed to the result of the selector function. This is quite powerful, because it means that even if we update the name of a todo, our component that only subscribes to the count via
useTodosCountwill not rerender. The count hasn't changed, so react-query can choose to not inform this observer about the update 🥳 (Please note that this is a bit simplified here and technically not entirely true - I will talk in more detail about render optimizations in Part 3).- 🟢 best optimizations
- 🟢 allows for partial subscriptions
- 🟡 structure can be different for every observer
- 🟡 structural sharing is performed twice (I will also talk about this in more detail in Part 3)
-
Data Transformation
[...]
2. In the render function
As advised in Part 1, if you create custom hooks, you can easily do transformations there:
/* render-transformation */ const fetchTodos = async (): Promise<Todos> => { const response = await axios.get('todos') return response.data } export const useTodosQuery = () => { const queryInfo = useQuery(['todos'], fetchTodos) return { ...queryInfo, data: queryInfo.data?.map((todo) => todo.name.toUpperCase()), } }As it stands, this will not only run every time your fetch function runs, but actually on every render (even those that do not involve data fetching). This is likely not a problem at all, but if it is, you can optimize with
useMemo. Be careful to define your dependencies as narrow as possible. data inside thequeryInfowill be referentially stable unless something really changed (in which case you want to recompute your transformation), but thequeryInfoitself will not. If you addqueryInfoas your dependency, the transformation will again run on every render:/* useMemo-dependencies */ export const useTodosQuery = () => { const queryInfo = useQuery(['todos'], fetchTodos) return { ...queryInfo, // 🚨 don't do this - the useMemo does nothing at all here! data: React.useMemo( () => queryInfo.data?.map((todo) => todo.name.toUpperCase()), [queryInfo] ), // ✅ correctly memoizes by queryInfo.data data: React.useMemo( () => queryInfo.data?.map((todo) => todo.name.toUpperCase()), [queryInfo.data] ), } }Especially if you have additional logic in your custom hook to combine with your data transformation, this is a good option. Be aware that data can be potentially undefined, so use optional chaining when working with it.
- 🟢 optimizable via useMemo
- 🟡 exact structure cannot be inspected in the devtools
- 🔴 a bit more convoluted syntax
- 🔴 data can be potentially undefined
-
-
jenweber.dev jenweber.dev
-
-
codesandbox.io codesandbox.io
-
-
www.dailymotion.com www.dailymotion.com
-
-
www.dailymotion.com www.dailymotion.com
-
-
vimeo.com vimeo.com
Tags
Annotators
URL
-
-
-
-
scicrunch.org scicrunch.org
-
-
csstracking.dev csstracking.dev
-
-
hn.algolia.com hn.algolia.com
-
-
lisperator.net lisperator.net
Tags
Annotators
URL
-
-
www.slightedgecoder.com www.slightedgecoder.com
-
css-tricks.com css-tricks.com
-
-
css-tricks.com css-tricks.com
Tags
Annotators
URL
-
-
-
-
www.w3.org www.w3.org
-
-
blog.jonudell.net blog.jonudell.net
-
www.sitepoint.com www.sitepoint.com
Tags
Annotators
URL
-
-
bionicjulia.com bionicjulia.com
-
mxb.dev mxb.dev
Tags
Annotators
URL
-
-
iipc.github.io iipc.github.io
-
Making a Memento
To create an archived version of the page that could be played back properly, I used the Internet Archive’s “Save” feature by going to this URL in my web browser:
…which created this snapshot:
From here, we can use
wgetto look at what gets played back:$ wget --server-response http://web.archive.org/web/20150709104019/http://iipc.github.io/warc-specifications/primers/web-archive-formats/hello-world.txt…giving:
HTTP/1.0 200 OK Server: Tengine/2.1.0 Date: Thu, 09 Jul 2015 10:41:38 GMT Content-Type: text/plain;charset=utf-8 Content-Length: 13 Set-Cookie: wayback_server=19; Domain=archive.org; Path=/; Expires=Sat, 08-Aug-15 10:41:38 GMT; Memento-Datetime: Thu, 09 Jul 2015 10:40:19 GMT Link: <http://iipc.github.io/warc-specifications/primers/web-archive-formats/hello-world.txt>; rel="original", <http://web.archive.org/web/timemap/link/http://iipc.github.io/warc-specifications/primers/web-archive-formats/hello-world.txt>; rel="timemap"; type="application/link-format", <http://web.archive.org/web/http://iipc.github.io/warc-specifications/primers/web-archive-formats/hello-world.txt>; rel="timegate", <http://web.archive.org/web/20150709104019/http://iipc.github.io/warc-specifications/primers/web-archive-formats/hello-world.txt>; rel="first last memento"; datetime="Thu, 09 Jul 2015 10:40:19 GMT" X-Archive-Orig-x-cache-hits: 0 X-Archive-Orig-x-served-by: cache-sjc3122-SJC X-Archive-Orig-cache-control: max-age=600 X-Archive-Orig-content-type: text/plain; charset=utf-8 X-Archive-Orig-server: GitHub.com X-Archive-Orig-age: 0 X-Archive-Orig-x-timer: S1436438419.302921,VS0,VE141 X-Archive-Orig-access-control-allow-origin: * X-Archive-Orig-last-modified: Wed, 08 Jul 2015 22:33:03 GMT X-Archive-Orig-expires: Thu, 09 Jul 2015 10:50:19 GMT X-Archive-Orig-accept-ranges: bytes X-Archive-Orig-vary: Accept-Encoding X-Archive-Orig-connection: close X-Archive-Orig-date: Thu, 09 Jul 2015 10:40:19 GMT X-Archive-Orig-via: 1.1 varnish X-Archive-Orig-content-length: 13 X-Archive-Orig-x-cache: MISS X-Archive-Wayback-Perf: {"IndexLoad":359,"IndexQueryTotal":359,"RobotsFetchTotal":1,"RobotsRedis":1,"RobotsTotal":1,"Total":371,"WArcResource":10} X-Archive-Playback: 1 X-Page-Cache: MISS -
Extracting a WARC record
Once we’ve identified the offset and length of a particular record (in this case, an offset of 1260 bytes and a length of 1085 bytes), we can snip out an individual record like this:
$ tail -c +1261 hello-world.warc | head -c 1085 -
Making the CDX
To generate a content index (CDX) file, we have at least two options. There’s JWATTools:
$ jwattools cdx hello-world.warc…(which created cdx.unsorted.out), or the
cdx-indexerfrom OpenWayback:$ cdx-indexer hello-world.warc > hello-world.warc.cdx…(which created hello-world.warc.cdx).
-
Making the WARC
To create a WARC, we used
wget:$ wget --warc-file hello-world http://iipc.github.io/warc-specifications/primers/web-archive-formats/hello-world.txt…which created the compressed hello-world.warc.gz file. These special block-compressed files are often used directly, but in this primer, we uncompress it so we can see what’s going on:
$ gunzip hello-world.warc.gz…leaving us with hello-world.warc.
-
-
stackoverflow.com stackoverflow.com
-
Add a
::afterwhich autofills the space. No need to pollute your HTML. Here is a codepen showing it: http://codepen.io/DanAndreasson/pen/ZQXLXj.grid { display: flex; flex-flow: row wrap; justify-content: space-between; } .grid::after { content: ""; flex: auto; }
-
-
javascript.info javascript.info
-
-
www.w3.org www.w3.org
-
<figure> 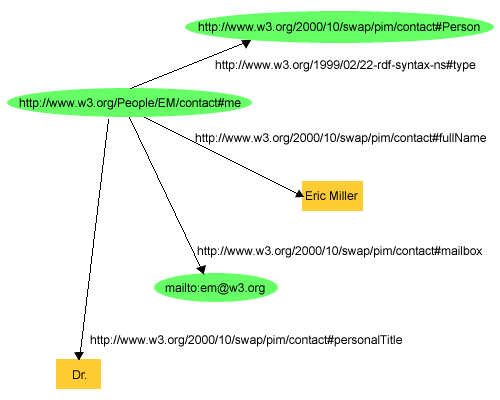 <caption>Figure 1: An RDF Graph Describing Eric Miller</caption>
</figure>
<caption>Figure 1: An RDF Graph Describing Eric Miller</caption>
</figure>
-
-
randomgeekery.org randomgeekery.org
-
webmention.herokuapp.com webmention.herokuapp.com
-
-
keithjgrant.com keithjgrant.com
-
-
🤔 So what?
Let’s review a typical Redux process for a fetch operation:
- Dispatch an Action
fetchSomething()from within the Component - This Action hits the Reducer which will update the store with something like
isLoading: true - The Component re-renders and displays a loader
- In the meanwhile the Action hits the Middleware which takes care of calling the API
- The API returns the response to the Middleware, which dispatches another Action:
fetchSomethingSucceeded()when succeeded, orfetchSomethingFailed()when failed - The succeeded/failed Action hits the Reducer, which updates the
store - Finally, the response is back to the Component (optionally through a memoized selector) which will show the data
It’s a long process for just a simple fetch, isn’t it?
- Dispatch an Action
-
-
www.flickr.com www.flickr.com
-
tkdodo.eu tkdodo.eu
-
www.crossref.org www.crossref.org
Tags
Annotators
URL
-
- Dec 2021
-
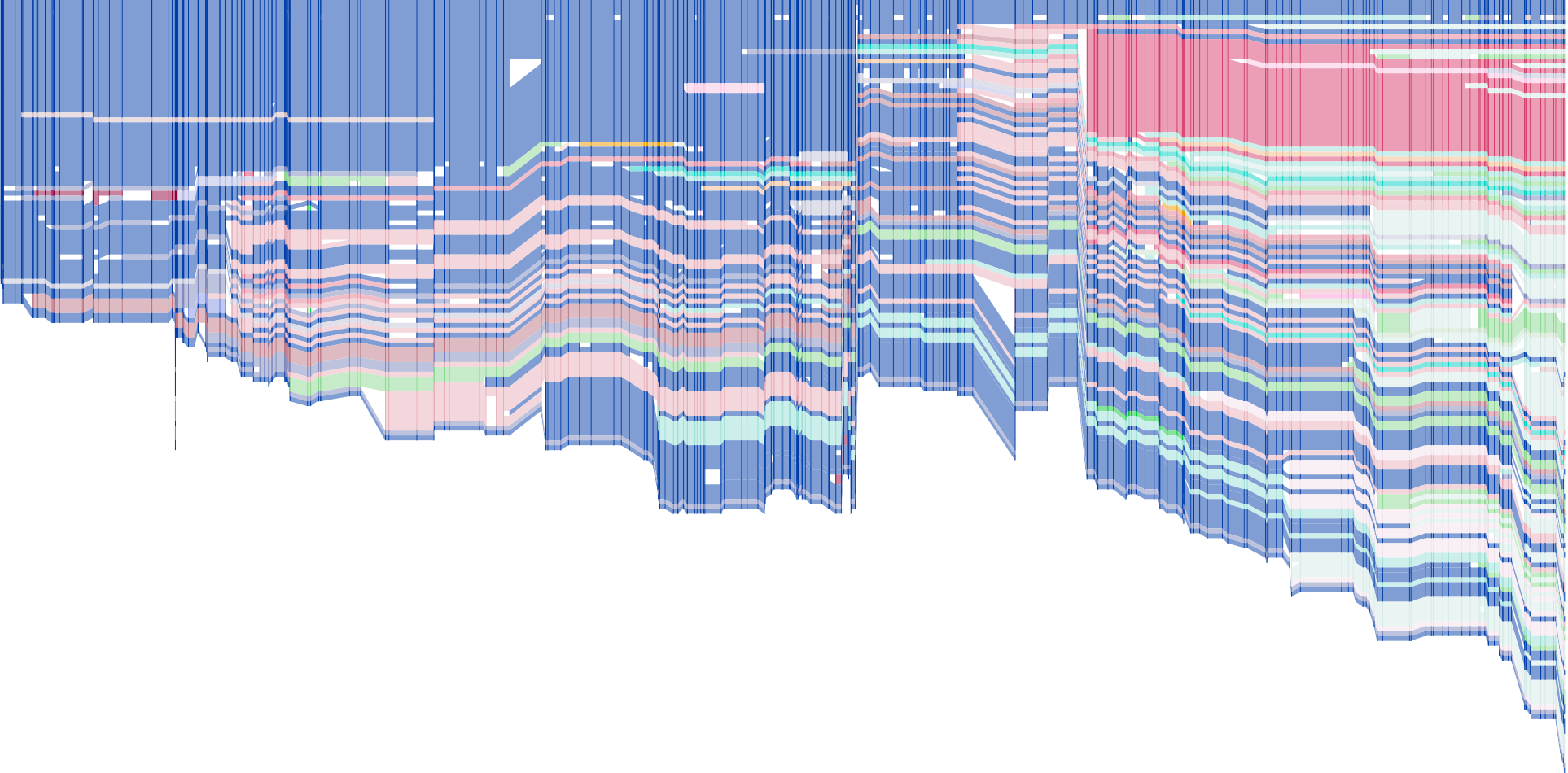
github.com github.com
-
This code creates a simple web site that queries Wikipedia to get the edit history of a page and renders it as a "history flow" in SVG.
<figure>  </figure>
</figure>
-
-
adotg.github.io adotg.github.io
-
Visualize the evolution of a file tracked by git.
Tags
Annotators
URL
-
-
yearsofnolight.medium.com yearsofnolight.medium.com
-
<figure>
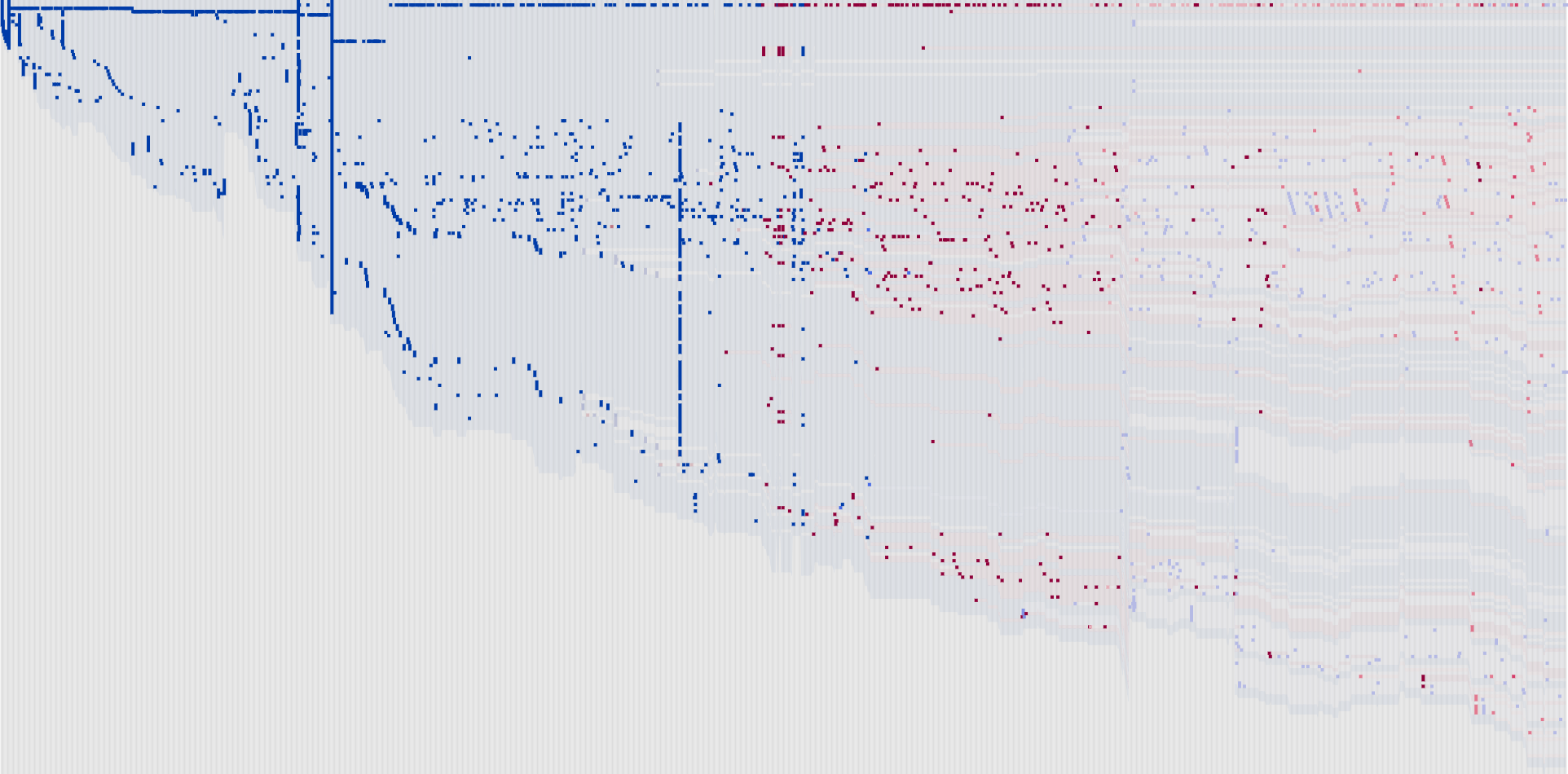
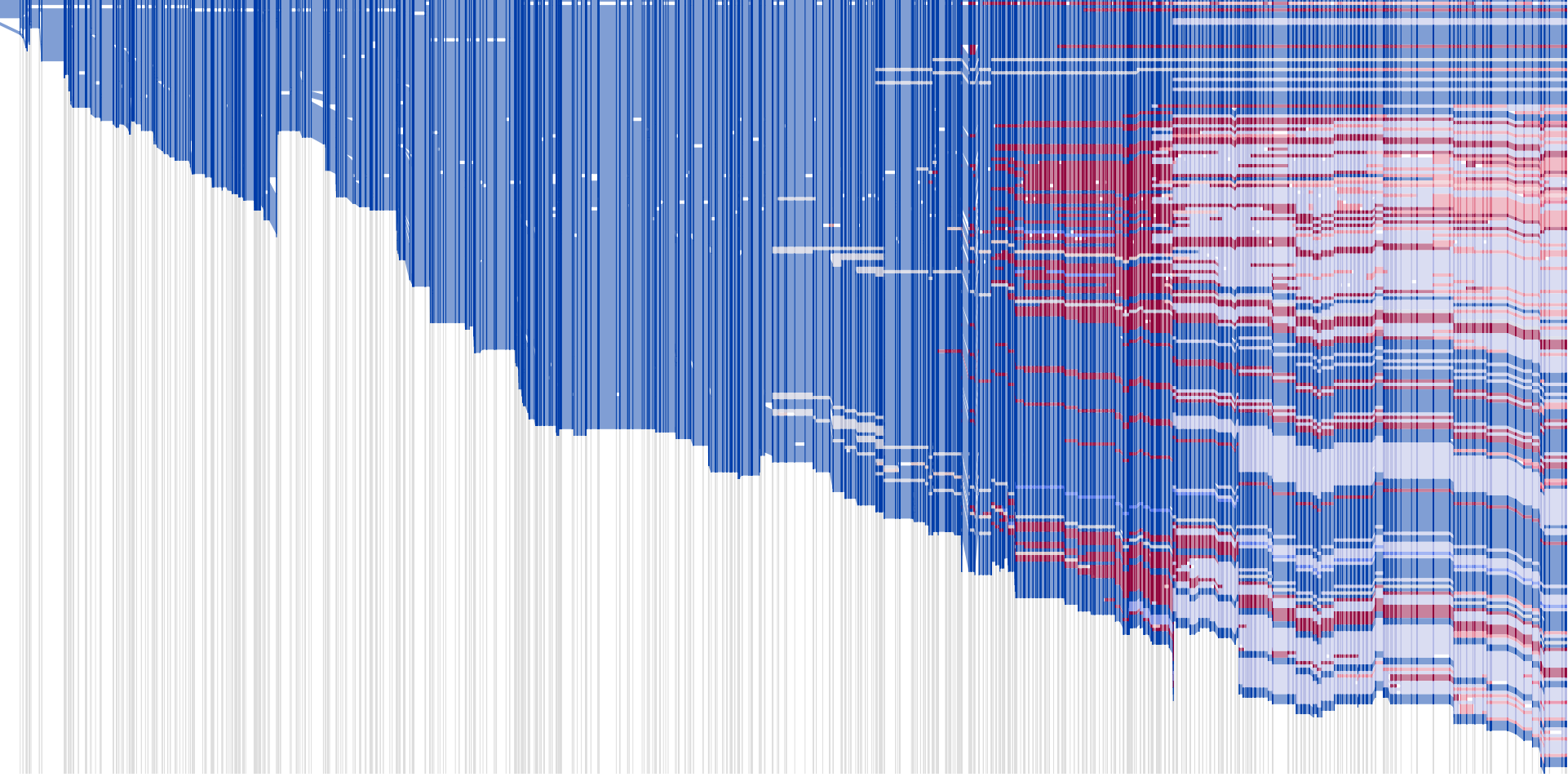 </figure/>
</figure/>
-
-
researchweb.watson.ibm.com researchweb.watson.ibm.com
-
visualizing the editing history of Wikipedia pages
<figure> 
 <figcaption>history flow is a tool for visualizing dynamic, evolving documents and the interactions of multiple collaborating authors. In its current implementation, history flow is being used to visualize the evolutionary history of wiki* pages on Wikipedia. </figcaption>
</figure>
<figcaption>history flow is a tool for visualizing dynamic, evolving documents and the interactions of multiple collaborating authors. In its current implementation, history flow is being used to visualize the evolutionary history of wiki* pages on Wikipedia. </figcaption>
</figure>
-
-
indieweb.org indieweb.org
-
A fragmention is an extension to URL syntax that links and cites a phrase within a document by using a URL fragment consisting of the phrase itself, including whitespace.
-
-
telegraph.p3k.io telegraph.p3k.io
-
-
boffosocko.com boffosocko.com
-
Marginalia
With Webmention support, one could architect a site to allow inline marginalia and highlighting similar to Medium.com’s relatively well-known functionality. With the clever use of URL fragments, which are well supported in major browsers, there are already examples of people who use Webmentions to display word-, sentence-, or paragraph-level marginalia on their sites. After all, aren’t inline annotations just a more targeted version of comments?
<figure> <figcaption>An inline annotation on the text “Hey Ev, what about mentions?” in which Medium began to roll out their @mention functionality.</figcaption>
</figure>
<figcaption>An inline annotation on the text “Hey Ev, what about mentions?” in which Medium began to roll out their @mention functionality.</figcaption>
</figure>
-
-
news.ycombinator.com news.ycombinator.com
-
Wayback Machine being broken in Firefox
Are you getting "Fail with status: 498 No Reason Phrase"? You might have your Referer header disabled.
If that's the case, you can fix it by going to about:config and setting network.http.sendRefererHeader to 2 (or pressing the reset button to the right).
-
-
tkdodo.eu tkdodo.eu
Tags
Annotators
URL
-
-
tkdodo.eu tkdodo.eu
Tags
Annotators
URL
-
-
tkdodo.eu tkdodo.eu
-
Increasing StaleTime
React Query comes with a default staleTime of zero. This means that every query will be immediately considered as stale, which means it will refetch when a new subscriber mounts or when the user refocuses the window. It is aimed to keep your data as up-to-date as necessary.
This goal overlaps a lot with WebSockets, which update your data in real-time. Why would I need to refetch at all if I just manually invalidated because the server just told me to do so via a dedicated message?
So if you update all your data via websockets anyways, consider setting a high
staleTime. In my example, I just usedInfinity. This means the data will be fetched initially via useQuery, and then always come from the cache. Refetching only happens via the explicit query invalidation.You can best achieve this by setting global query defaults when creating the QueryClient:
const queryClient = new QueryClient({ defaultOptions: { queries: { staleTime: Infinity, }, }, })
-
-
codesandbox.io codesandbox.io
Tags
Annotators
URL
-
-
thewebdev.info thewebdev.info
-
react-query.tanstack.com react-query.tanstack.com
-
blog.theodo.com blog.theodo.com
-
webpack.js.org webpack.js.org
-
Web Workers
As of webpack 5, you can use Web Workers without
worker-loader.Syntax
new Worker(new URL('./worker.js', import.meta.url));
Tags
Annotators
URL
-
-
web.dev web.dev
-
-
blog.openreplay.com blog.openreplay.com
-
Instant Dates and Times
Temporal.Instantreturns an object representing a date and time to the nearest nanosecond according to an ISO 8601 formatted string:
-
-
github.com github.com
-
// main.js const { RemoteReadableStream, RemoteWritableStream } = RemoteWebStreams; (async () => { const worker = new Worker('./worker.js'); // create a stream to send the input to the worker const { writable, readablePort } = new RemoteWritableStream(); // create a stream to receive the output from the worker const { readable, writablePort } = new RemoteReadableStream(); // transfer the other ends to the worker worker.postMessage({ readablePort, writablePort }, [readablePort, writablePort]); const response = await fetch('./some-data.txt'); await response.body // send the downloaded data to the worker // and receive the results back .pipeThrough({ readable, writable }) // show the results as they come in .pipeTo(new WritableStream({ write(chunk) { const results = document.getElementById('results'); results.appendChild(document.createTextNode(chunk)); // tadaa! } })); })();// worker.js const { fromReadablePort, fromWritablePort } = RemoteWebStreams; self.onmessage = async (event) => { // create the input and output streams from the transferred ports const { readablePort, writablePort } = event.data; const readable = fromReadablePort(readablePort); const writable = fromWritablePort(writablePort); // process data await readable .pipeThrough(new TransformStream({ transform(chunk, controller) { controller.enqueue(process(chunk)); // do the actual work } })) .pipeTo(writable); // send the results back to main thread };
-
-
solidproject.org solidproject.org
Tags
Annotators
URL
-
-
stackoverflow.com stackoverflow.com
-
What you're trying to do is known as the "Application Shell" architectural pattern.
The trick is to have your service worker's
fetchhandler check to see whether an incoming request is a navigation (event.request.mode === 'navigate'), and if so, respond with the cached App Shell HTML (which sounds like/index.htmlin your case).A generic way of doing this would be:
self.addEventListener('fetch', (event) => { if (event.request.mode === 'navigate') { event.respondWith(caches.match('/index.html')); } else { // Your other response logic goes here. } });This will cause your service worker to behave in a similar fashion to how you're web server is already configured.
-
-
hypothes.is hypothes.is
Tags
Annotators
URL
-
-
metro-parisien.mrsy.fr metro-parisien.mrsy.fr
Tags
Annotators
URL
-
-
stackedit.io stackedit.io
-
-
localhost:4000 localhost:4000
-
import warc from StringIO import StringIO from httplib import HTTPResponse class FakeSocket(): def __init__(self, response_str): self._file = StringIO(response_str) def makefile(self, *args, **kwargs): return self._file for record in warc.open("eada.warc.gz"): if record.type == "response": resp = HTTPResponse(FakeSocket(record.payload.read())) resp.begin() if resp.getheader("content-type") == "text/html": print record['WARC-Target-URI']I sorted the output and came up with a nice list of URLs for the website. Here is a brief snippet:
http://mith.umd.edu/eada/gateway/winslow.php http://mith.umd.edu/eada/gateway/winthrop.php http://mith.umd.edu/eada/gateway/witchcraft.php http://mith.umd.edu/eada/gateway/wood.php http://mith.umd.edu/eada/gateway/woolman.php http://mith.umd.edu/eada/gateway/yeardley.php http://mith.umd.edu/eada/guesteditors.php http://mith.umd.edu/eada/html/display.php?docs=acrelius_founding.xml&action=show http://mith.umd.edu/eada/html/display.php?docs=alsop_character.xml&action=show http://mith.umd.edu/eada/html/display.php?docs=arabic.xml&action=show http://mith.umd.edu/eada/html/display.php?docs=ashbridge_account.xml&action=show http://mith.umd.edu/eada/html/display.php?docs=banneker_letter.xml&action=show http://mith.umd.edu/eada/html/display.php?docs=barlow_anarchiad.xml&action=show http://mith.umd.edu/eada/html/display.php?docs=barlow_conspiracy.xml&action=show http://mith.umd.edu/eada/html/display.php?docs=barlow_vision.xml&action=show http://mith.umd.edu/eada/html/display.php?docs=barlowe_voyage.xml&action=show
Tags
Annotators
URL
-

