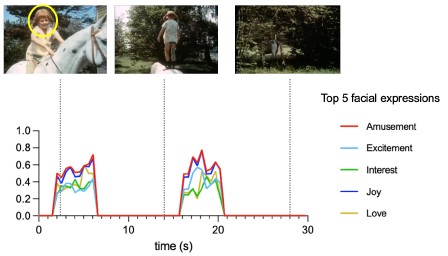
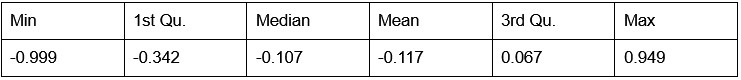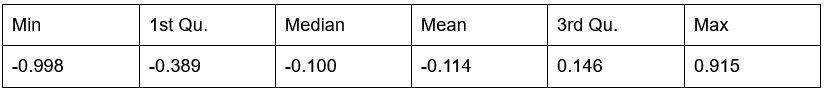"This implies that a sequence of uracils codes for phenylalanine, and our work suggests that it is probably a triplet of uracils." This was only a hypothesis when this paper was written and published, and now it is recognized as a biological fact. It's really crazy to see how quickly science can develop within just a few decades.
- Jan 2026
-
mssu.blackboard.com mssu.blackboard.comFile1
-
-
-
Author response:
The following is the authors’ response to the latest reviews:
"One remaining question is the interpretation of matching variants with very low stable posterior probabilities (~0), which the authors have analyzed in detail but without fully conclusive findings. I agree with the authors that this event is relatively rare and the current sample size is limited but this might be something to keep in mind for future studies."
Fine-mapping stability – on matching variants with very low stable posterior probability
We thank Reviewer 2 for encouraging us to think more about how low stable posterior probability matching variants can be interpreted. We describe a few plausible interpretations, even though – as Reviewer 2 and we have both acknowledged – our present experiments do not point to a clear and conclusive account.
One explanation is that the locus captured by the variant might not be well-resolved, in the sense that many correlated variants exist around the locus. Thus, the variant itself is unlikely causal, but the set of variants in high LD with it may contain the true causal variant, or it's possible that the causal variant itself was not sequenced but lies in that locus. A comparison of LD patterns across ancestries at the locus would be helpful here.
Another explanation rests on the following observation. For a variant to be matching between top and stable PICS and to also have very small stable PP, it has to have the largest PP after residualization on the ALL slice but also have positive PP with gene expression on many other slices. In other words, failing to control for potential confounders shrinks the PP. If one assumes that the matching variant is truly causal, then our observation points to an example of negative confounding (aka suppressor effect). This can occur when the confounders (PCs) are correlated with allele dosage at the causal variant in a different direction than their correlation with gene expression, so that the crude association between unresidualized gene expression and causal variant allele dosage is biased toward 0.
Although our present study does not allow us to systematically confirm either interpretation – since we found that matching variants were depleted in causal variants in our simulations, violating the second argument, but we also found functional enrichment in analyses of GEUVADIS data though only 17 matching variants with low stable PP were reported – we believe a larger-scale study using larger cohort sizes (at least 1000 individuals per ancestry) and many more simulations (to increase yield of such cases) would be insightful.
———
The following is the authors’ response to the original reviews:
Reviewer #1:
Major comments:
(1) It would be interesting to see how much fine-mapping stability can improve the fine-mapping results in cross-population. One can simulate data using true genotype data and quantify the amount the fine-mapping methods improve utilizing the stability idea.
We agree, and have performed simulation studies where we assume that causal variants are shared across populations. Specifically, by mirroring the simulation approach described in Wang et al. (2020), we generated 2,400 synthetic gene expression phenotypes across 22 autosomes, using GEUVADIS gene expression metadata (i.e., gene transcription start site) to ensure largely cis expression phenotypes were simulated. We additionally generated 1,440 synthetic gene expression phenotypes that incorporate environmental heterogeneity, to motivate our pursuit of fine-mapping stability in the first place (see Response to Reviewer 2, Comment 6). These are described in Results section “Simulation study”:
We evaluated the performance of the PICS algorithm, specifically comparing the approach incorporating stability guidance against the residualization approach that is more commonly used — similar to our application to the real GEUVADIS data. We additionally investigated two ways of “combining” the residualization and stability guidance approaches: (1) running stability-guided PICS on residualized phenotypes; (2) prioritizing matching variants returned by both approaches. See Response to Reviewer 2, Comment 5.
(2) I would be very interested to see how other fine-mapping methods (FINEMAP, SuSiE, and CAVIAR) perform via the stability idea.
Thank you for this valuable comment. We ran SuSiE on the same set of simulated datasets. Specifically, we ran a version that uses residualized phenotypes (supposedly removing the effects of population structure), and also a version that incorporates stability. The second version is similar to how we incorporate stability in PICS. We investigated the performance of Stable SuSiE in a similar manner to our investigation of PICS. First we compared the performance relative to SuSiE that was run on residualized phenotypes. Motivated by our finding in PICS that prioritizing matching variants improves causal variant recovery, we did the same analysis for SuSiE. This analysis is described in Results section “Stability guidance improves causal variant recovery in SuSiE.”
We reported overall matching frequencies and causal variant recovery rates of top and stable variants for SuSiE in Figures 2C&D.
Frequencies with which Stable and Top SuSiE variants match, stratified by the simulation parameters, are summarized in Supplementary File 2C (reproduced for convenience in Response to Reviewer 2, Comment 3). Causal variant recovery rates split by the number of causal variants simulated, and stratified by both signal-to-noise ratio and the number of credible sets included, are reported in Figure 2—figure supplements 16-18. We reproduce Figure 2—figure supplement 18 (three causal variants scenario) below for convenience. Analogous recovery rates for matching versus non-matching top or stable variants are reported in Figure 2—figure supplements 19, 21 and 23.
(3) I am a little bit concerned about the PICS's assumption about one causal variant. The authors mentioned this assumption as one of their method limitations. However, given the utility of existing fine-mapping methods (FINEMAP and SuSiE), it is worth exploring this domain.
Thank you for raising this fair concern. We explored this domain, by considering simulations that include two and three causal variants (see Response to Reviewer 2, Comment 3). We looked at how well PICS recovers causal variants, and found that each potential set largely does not contain more than one causal variant (Figure 2—figure supplements 20 and 22). This can be explained by the fact that PICS potential sets are constructed from variants with a minimum linkage disequilibrium to a focal variant. On the other hand, in SuSiE, we observed multiple causal variants appearing in lower credible sets when applying stability guidance (Figure 2—figure supplements 21 and 23). A more extensive study involving more fine-mapping methods and metrics specific to violation of the one causal variant assumption could be pursued in future work.
Reviewer #2:
Aw et al. presents a new stability-guided fine-mapping method by extending the previously proposed PICS method. They applied their stability-based method to fine-map cis-eQTLs in the GEUVADIS dataset and compared it against what they call residualization-based method. They evaluated the performance of the proposed method using publicly available functional annotations and claimed the variants identified by their proposed stability-based method are more enriched for these functional annotations.
While the reviewer acknowledges the contribution of the present work, there are a couple of major concerns as described below.
Major:
(1) It is critical to evaluate the proposed method in simulation settings, where we know which variants are truly causal. While I acknowledge their empirical approach using the functional annotations, a more unbiased, comprehensive evaluation in simulations would be necessary to assess its performance against the existing methods.
Thank you for this point. We agree. We have performed a simulation study where we assume that causal variants are shared across populations (see response to Reviewer 1, Comment 1). Specifically, by mirroring the simulation approach described in Wang et al. (2020), we generated 2,400 synthetic gene expression phenotypes across 22 autosomes, using GEUVADIS gene expression metadata (i.e., gene transcription start site) to ensure cis expression phenotypes were simulated.
(2) Also, simulations would be required to assess how the method is sensitive to different parameters, e.g., LD threshold, resampling number, or number of potential sets.
Thank you for raising this point. The underlying PICS algorithm was not proposed by us, so we followed the default parameters set (LD threshold, r<sup>2</sup> \= 0.5; see Taylor et al., 2021 Bioinformatics) to focus on how stability considerations will impact the existing fine-mapping algorithm. We attempted to derive the asymptotic joint distribution of the p-values, but it was too difficult. Hence, we used 500 permutations because such a large number would allow large-sample asymptotics to kick in. However, following your critical suggestion we varied the number of potential sets in our analyses of simulated data. We briefly mention this in the Results.
“In the Supplement, we also describe findings from investigations into the impact of including more potential sets on matching frequency and causal variant recovery…”
A detailed write-up is provided in Supplementary File 1 Section S2 (p.2):
“The number of credible or potential sets is a parameter in many fine-mapping algorithms. Focusing on stability-guided approaches, we consider how including more potential sets for stable fine-mapping algorithms affects both causal variant recovery and matching frequency in simulations…
Causal variant recovery. We investigate both Stable PICS and Stable SuSiE. Focusing first on simulations with one causal variant, we observe a modest gain in causal variant recovery for both Stable PICS and Stable SuSiE, most noticeably when the number of sets was increased from 1 to 2 under the lowest signal-to-noise ratio setting…”
We observed that increasing the number of potential sets helps with recovering causal variants for Stable PICS (Figure 2—figure supplements 13-15). This observation also accounts for the comparable power that Stable PICS has with SuSiE in simulations with low signal-to-noise ratio (SNR), when we increase the number of credible sets or potential sets (Figure 2—figure supplements 10-12).
(3) Given the previous studies have identified multiple putative causal variants in both GWAS and eQTL, I think it's better to model multiple causal variants in any modern fine-mapping methods. At least, a simulation to assess its impact would be appreciated.
We agree. In our simulations we considered up to three causal variants in cis, and evaluated how well the top three Potential Sets recovered all causal variants (Figure 2—figure supplements 13-15; Figure 2—figure supplement 15). We also reported the frequency of variant matches between Top and Stable PICS stratified by the number of causal variants simulated in Supplementary File 2B and 2C. Note Supplementary File 2C is for results from SuSiE fine-mapping; see Response to Reviewer 1, Comment 2.
Supplementary File 2B. Frequencies with which Stable and Top PICS have matching variants for the same potential set. For each SNR/ “No. Causal Variants” scenario, the number of matching variants is reported in parentheses.
Supplementary File 2C. Frequencies with which Stable and Top SuSiE have matching variants for the same credible set. For each SNR/ “No. Causal Variants” scenario, the number of matching variants is reported in parentheses.
(4) Relatedly, I wonder what fraction of non-matching variants are due to the lack of multiple causal variant modeling.
PICS handles multiple causal variants by including more potential sets to return, owing to the important caveat that causal variants in high LD cannot be statistically distinguished. For example, if one believes there are three causal variants that are not too tightly linked, one could make PICS return three potential sets rather than just one. To answer the question using our simulation study, we subsetted our results to just scenarios where the top and stable variants do not match. This mimics the exact scenario of having modeled multiple causal variants but still not yielding matching variants, so we can investigate whether these non-matching variants are in fact enriched in the true causal variants.
Because we expect causal variants to appear in some potential set, we specifically considered whether these non-matching causal variants might match along different potential sets across the different methods. In other words, we compared the stable variant with the top variant from another potential set for the other approach (e.g., Stable PICS Potential Set 1 variant vs Top PICS Potential Set 2 variant). First, we computed the frequency with which such pairs of variants match. A high frequency would demonstrate that, even if the corresponding potential sets do not have a variant match, there could still be a match between non-corresponding potential sets across the two approaches, which shows that multiple causal variant modeling boosts identification of matching variants between both approaches — regardless of whether the matching variant is in fact causal.
Low frequencies were observed. For example, when restricting to simulations where Top and Stable PICS Potential Set 1 variants did not match, about 2-3% of variants matched between the Potential Set 1 variant in Stable PICS and Potential Sets 2 and 3 variants in Top PICS; or between the Potential Set 1 variant in Top PICS and Potential Sets 2 and 3 variants in Stable PICS (Supplementary File 2D). When looking at non-matching Potential Set 2 or Potential Set 3 variants, we do see an increase in matching frequencies (between 10-20%) between Potential Set 2 variants and other potential set variants between the different approaches. However, these percentages are still small compared to the matching frequencies we observed between corresponding potential sets (e.g., for simulations with one causal variant this was 70-90% between Top and Stable PICS Potential Set 1, and for simulations with two and three causal variants this was 55-78% and 57-79% respectively).
We next checked whether these “off-diagonal” matching variants corresponded to the true causal variants simulated. Here we find that the causal variant recovery rate is mostly less than the corresponding rate for diagonally matching variants, which together with the low matching frequency suggests that the enrichment of causal variants of “off-diagonal” matching variants is much weaker than in the diagonally matching approach. In other words, the fraction of non-matching (causal) variants due to the lack of multiple causal variant modeling is low.
We discuss these findings in Supplementary File 1 Section S2 (bottom of p.2).
(5) I wonder if you can combine the stability-based and the residualization-based approach, i.e., using the residualized phenotypes for the stability-based approach. Would that further improve the accuracy or not?
This is a good idea, thank you for suggesting it. We pursued this combined approach on simulated gene expression phenotypes, but did not observe significant gains in causal variant recovery (Figure 2B; Figure 2—figure supplements 2, 13 and 15). We reported this Results “Searching for matching variants between Top PICS and Stable PICS improves causal variant Recovery.”
“We thus explore ways to combine the residualization and stability-driven approaches, by considering (i) combining them into a single fine-mapping algorithm (we call the resulting procedure Combined PICS); and (ii) prioritizing matching variants between the two algorithms. Comparing the performance of Combined PICS against both Top and Stable PICS, however, we find no significant difference in its ability to recover causal variants (Figure 2B)...”
However, we also confirmed in our simulations that prioritizing matching variants between the two approaches led to gains in causal variant recovery (Figure 2D; Figure 2—figure supplements 4, 19, 20 and 22). We reported this Results “Searching for matching variants between Top PICS and Stable PICS improves causal variant Recovery.”
“On the other hand, matching variants between Top and Stable PICS are significantly more likely to be causal. Across all simulations, a matching variant in Potential Set 1 is 2.5X as likely to be causal than either a non-matching top or stable variant (Figure 2D) — a result that was qualitatively consistent even when we stratified simulations by SNR and number of causal variants simulated (Figure 2—figure supplements 19, 20 and 22)...”
This finding is consistent with our analysis of real GEUVADIS gene expression data, where we reported larger functional significance of matching variants relative to non-matching variants returned by either Top of Stable PICS.
(6) The authors state that confounding in cohorts with diverse ancestries poses potential difficulties in identifying the correct causal variants. However, I don't see that they directly address whether the stability approach is mitigating this. It is hard to say whether the stability approach is helping beyond what simpler post-hoc QC (e.g., thresholding) can do.
Thank you for raising this fair point. Here is a model we have in mind. Gene expression phenotypes (Y) can be explained by both genotypic effects (G, as in genotypic allelic dosage) and the environment (E): Y = G + E. However, both G and E depend on ancestry (A), so that Y = G|A+E|A. Suppose that the causal variants are shared across ancestries, so that (G|A=a)=G for all ancestries a. Suppose however that environments are heterogeneous by ancestry: (E|A=a) = e(a) for some function e that depends non-trivially on a. This would violate the exchangeability of exogenous E in the full sample, but by performing fine-mapping on each ancestry stratum, the exchangeability of exogenous E is preserved. This provides theoretical justification for the stability approach.
We next turned to simulations, where we investigated 1,440 simulated gene expression phenotypes capturing various ways in which ancestry induces heterogeneity in the exogenous E variable (simulation details in Lines 576-610 of Materials and Methods). We ran Stable PICS, as well as a version of PICS that did not residualize phenotypes or apply the stability principle. We observed that (i) causal variant recovery performance was not significantly different between the two approaches (Figure 2—figure supplements 24-32); but (ii) disagreement between the approaches can be considerable, especially when the signal-to-noise ratio is low (Supplementary File 2A). For example, in a set of simulations with three causal variants, with SNR = 0.11 and E heterogeneous by ancestry by letting E be drawn from N(2σ,σ<sup>2</sup>) for only GBR individuals (rest are N(0,σ<sup>2</sup>)), there was disagreement between Potential Set 1 and 2 variants in 25% of simulations — though recovery rates were similar (Probability of recovering at least one causal variant: 75% for Plain PICS and 80% for Stable PICS). These points suggest that confounding in cohorts can reduce power in methods not adjusting or accounting for ancestral heterogeneity, but can be remedied by approaches that do so. We report this analysis in Results “Simulations justify exploration of stability guidance”
In the current version of our work, we have evaluated, using both simulations and empirical evidence, different ways to combine approaches to boost causal variant recovery. Our simulation study shows that prioritizing matching variants across multiple methods improves causal variant recovery. On GEUVADIS data, where we might not know which variants are causal, we already demonstrated that matching variants are enriched for functional annotations. Therefore, our analyses justify that the adverse consequence of confounding on reducing fine-mapping accuracy can be mitigated by prioritizing matching variants between algorithms including those that account for stability.
(7) For non-matching variants, I wonder what the difference of posterior probabilities is between the stable and top variants in each method. If the difference is small, maybe it is due to noise rather than signal.
We have reported differences in posterior probabilities returned by Stable and Top PICS for GEUVADIS data; see Figure 3—figure supplement 1. For completeness, we compute the differences in posterior probabilities and summarize these differences both as histograms and as numerical summary statistics.
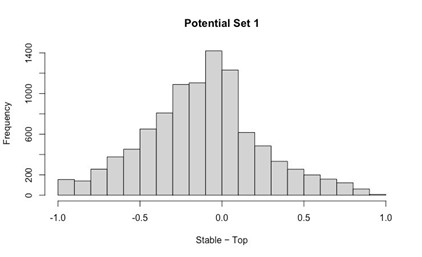
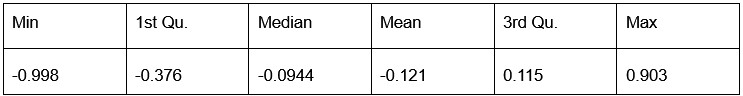
Potential Set 1
- Number of non-matching variants = 9,921
- Table of Summary Statistics of (Stable Posterior Probability – Top Posterior Probability)
Author response table 1.
- Histogram of (Stable Posterior Probability – Top Posterior Probability)
Author response image 1.
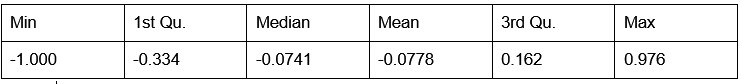
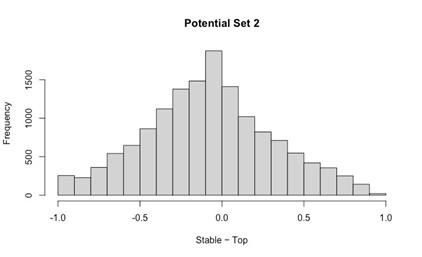
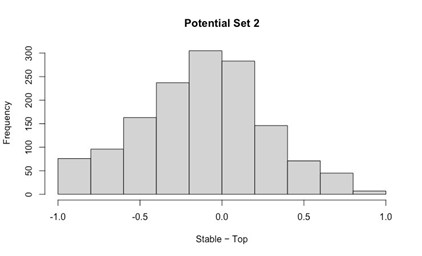
Potential Set 2
- Number of non-matching variants = 14,454
- Table of Summary Statistics of (Stable Posterior Probability – Top Posterior Probability)
Author response table 2.
- Histogram of (Stable Posterior Probability – Top Posterior Probability)
Author response image 2.
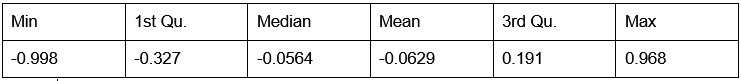
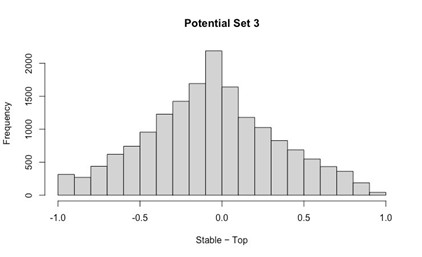
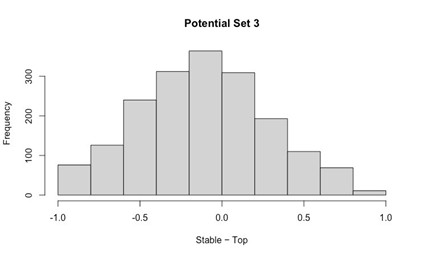
Potential Set 3
- Number of non-matching variants = 16,814
- Table of Summary Statistics of (Stable Posterior Probability – Top Posterior Probability)
Author response table 3.
- Histogram of (Stable Posterior Probability – Top Posterior Probability)
Author response image 3.
We also compared the difference in posterior probabilities between non-matching variants returned by Stable PICS and Top PICS for our 2,400 simulated gene expression phenotypes. Focusing on just Potential Set 1 variants, we find two equally likely scenarios, as demonstrated by two distinct clusters of points in a “posterior probability-posterior probability” plot. The first is, as pointed out, a small difference in posterior probability (points lying close to y=x). The second, however, reveals stable variants with very small posterior probability (of order 4 x 10<sup>–5</sup> to 0.05) but with a non-matching top variant taking on posterior probability well distributed along [0,1]. Moving down to Potential Sets 2 and 3, the distribution of pairs of posterior probabilities appears less clustered, indicating less tendency for posterior probability differences to be small ( Figure 2—figure supplement 8).
Here are the histograms and numerical summary statistics.
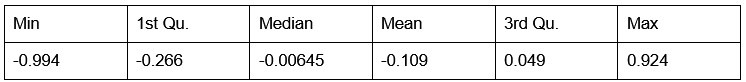
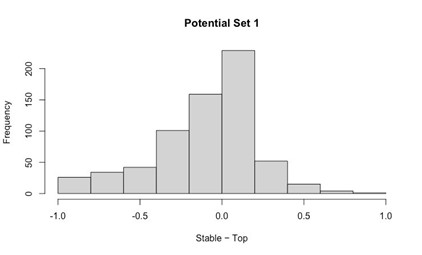
Potential Set 1
- Number of non-matching variants = 663 (out of 2,400)
- Table of Summary Statistics of (Stable Posterior Probability – Top Posterior Probability)
Author response table 4.
- Histogram of (Stable Posterior Probability – Top Posterior Probability)
Author response image 4.
Potential Set 2
Number of non-matching variants = 1,429 (out of 2,400)
- Table of Summary Statistics of (Stable Posterior Probability – Top Posterior Probability)
Author response table 5.
- Histogram of (Stable Posterior Probability – Top Posterior Probability)
Author response image 5.
Potential Set 3
- Number of non-matching variants = 1,810 (out of 2,400)
- Table of Summary Statistics of (Stable Posterior Probability – Top Posterior Probability)
Author response table 6.
- Histogram of (Stable Posterior Probability – Top Posterior Probability)
Author response image 6.
(8) It's a bit surprising that you observed matching variants with (stable) posterior probability ~ 0 (SFig. 1). What are the interpretations for these variants? Do you observe functional enrichment even for low posterior probability matching variants?
Thank you for this question. We have performed a thorough analysis of matching variants with very low stable posterior probability, which we define as having a posterior probability < 0.01 (Supplementary File 1 Section S11). Here, we briefly summarize the analysis and key findings.
Analysis
First, such variants occur very rarely — only 8 across all three potential sets in simulations, and 17 across all three potential sets for GEUVADIS (the latter variants are listed in Supplementary 2E). We begin interpreting these variants by looking at allele frequency heterogeneity by ancestry, support size — defined as the number of variants with positive posterior probability in the ALL slice* — and the number of slices including the stable variant (i.e., the stable variant reported positive posterior probability for the slice).
*Note that the stable variant posterior probability need not be at least 1/(Support Size). This is because the algorithm may have picked a SNP that has a lower posterior probability in the ALL slice (i.e., not the top variant) but happens to appear in the most number of other slices (i.e., a stable variant).
For variants arising from simulations, because we know the true causal variants, we check if these variants are causal. For GEUVADIS fine-mapped variants, we rely on functional annotations to compare their relative enrichment against other matching variants that did not have very low stable posterior probability.
Findings
While we caution against generalizing from observations reported here, which are based on very small sample sizes, we noticed the following. In simulations, matching variants with very low stable posterior probability are largely depleted in causal variants, although factors such as the number of slices including the stable variant may still be useful. In GEUVADIS, however, these variants can still be functionally enriched. We reported three examples in Supplementary File 1 Section S11 (pp. 8-9 of Supplement), where the variants were enriched in either VEP or biologically interpretable functional annotations, and were also reported in earlier studies. We partially reproduce our report below for convenience.
“However, we occasionally found variants that stand out for having large functional annotation scores. We list one below for each potential set.
- Potential Set 1 reported the variant rs12224894 from fine-mapping ENSG00000255284.1 (accession code AP006621.3) in Chromosome 11. This variant stood out for lying in the promoter flanking region of multiple cell types and being relatively enriched for GC content with a 75bp flanking region. This variant has been reported as a cis eQTL for AP006632 (using whole blood gene expression, rather than lymphoblastoid cell line gene expression in this study) in a clinical trial study of patients with systemic lupus erythematosus (Davenport et al., 2018). Its nearest gene is GATD1, a ubiquitously expressed gene that codes for a protein and is predicted to regulate enzymatic and catabolic activity. This variant appeared in all 6 slices, with a moderate support size of 23.
- Potential Set 2 reported the variant rs9912201 from fine-mapping ENSG00000108592.9 (mapped to FTSJ3) in Chromosome 17. Its FIRE score is 0.976, which is close to the maximum FIRE score reported across all Potential Set 2 matching variants. This variant has been reported as a SNP in high LD to a GWAS hit SNP rs7223966 in a pan-cancer study (Gong et al., 2018). This variant appeared in all 6 slices, with a moderate support size of 32.
- Potential Set 3 reported the variant rs625750 from fine-mapping ENSG00000254614.1 (mapped to CAPN1-AS1, an RNA gene) in Chromosome 11. Its FIRE score is 0.971 and its B statistic is 0.405 (region under selection), which lie at the extreme quantiles of the distributions of these scores for Potential Set 3 matching variants with stable posterior probability at least 0.01. Its associated mutation has been predicted to affect transcription factor binding, as computed using several position weight matrices (Kheradpour and Kellis, 2014). This variant appeared in just 3 slices, possibly owing to the considerable allele frequency difference between ancestries (maximum AF difference = 0.22). However, it has a small support size of 4 and a moderately high Top PICS posterior probability of 0.64.
To summarize, our analysis of GEUVADIS fine-mapped variants demonstrates that matching variants with very low stable posterior probability could still be functionally important, even for lower potential sets, conditional on supportive scores in interpretable features such as the number of slices containing the stable variant and the posterior probability support size…”
-
-
iowastatedaily.com iowastatedaily.com
-
“Digital badges are a great opportunity for Iowa State students to explore different avenues to build career-readiness skills,” Hageman said. “It’s not just a badge, it’s the work you put into earning it that stands out.”
Student quote
-
-
www.windowscentral.com www.windowscentral.com
-
Hundreds of millions of us have already given away ownership over music, TV shows, and movies to cloud companies like Spotify and Netflix — both of which run on Amazon Web Services. Cloud gaming products like Amazon Luna, NVIDIA GeForce Now, and Xbox Cloud Gaming are all seeing steady growth, too — but it's not just about these niche scenarios.
fair point, we do need to bring media home again. i've made the switch in books early last year. Music up next.
-
-
Local file Local file
-
olitics and the New Machine
The core argument
The essay argues that polling has become less reliable at the same time that it has become more powerful, and that this combination distorts democratic politics.
Polls:
increasingly fail to accurately measure public opinion
yet increasingly determine who gets attention, legitimacy, money, debate access, and media coverage
How Trump fits in
The piece opens with Donald Trump claiming he has no pollster and doesn’t tailor his message to polls. Lepore calls this disingenuous:
Trump may not have had a traditional campaign pollster
but his rise depended heavily on polls for visibility and validation
polls got him into debates, dictated stage placement, and fueled media coverage
So Trump is described as “a creature of the Sea of Polls,” not above it
Why modern polls are broken
The article explains in detail why polling has deteriorated:
- People don’t answer anymore
Response rates used to be 60–90%
Now they’re often in the single digits
Most Americans refuse poll calls, creating non-response bias
- Technology & law made it worse
Fewer landlines
Cell-phone autodialing is illegal
Internet polls are self-selected and skew younger and more liberal
Mixed-method polling still doesn’t work well
- Samples are tiny and fragile
National election polls often rely on ~1,000–2,000 people
Statistical “weighting” tries to fix bias, but the lower the response rate, the shakier the results
Why polls now matter more than ever
Despite being unreliable, polls are used to:
decide who qualifies for debates
determine media attention
shape fundraising and momentum
create “winners” and “losers” long before anyone votes
Fox News using polls to select debate participants is presented as a major example of polling replacing democratic processes.
Historical background
The essay gives a history of polling:
Early “straw polls” by newspapers
The rise of George Gallup in the 1930s
Polling claimed to represent “the will of the people” scientifically
But:
Early polls systematically excluded Black Americans, the poor, and the disenfranchised
Polling mirrored and amplified existing inequalities
What was presented as “public opinion” was often the opinion of a privileged subset
Deeper philosophical critique
Lepore raises a fundamental question:
What if measuring public opinion isn’t good for democracy at all?
Key ideas:
Polls treat public opinion as the sum of individual answers, ignoring how opinions are formed socially
Polls can create opinion rather than measure it
Constant polling shifts politics from deliberation and leadership to reacting to numbers
Bottom line
The piece isn’t just saying “polls are inaccurate.”
It’s saying:
Polls shape reality instead of describing it
They weaken representative democracy
They reward spectacle, momentum, and media attention over governance
And they increasingly substitute statistical artifacts for actual voting
-
-
publish.obsidian.md publish.obsidian.md
-
Weapons made of obsidian, jewelry crafted from jade
Obsidian often is known for protecting you against negative energies and psychic attacks in some religions, besides it being a sharp volcanic glass that can easily pierce victims of its wrath. It's also good for grounding you, or in other words, just keeping you in check with reality. As for jade, it can have several benefits in some cultures depending on the color. For example, green jade (usually seen in bangle form in countries like Vietnam and Myanmar/Burma) can represent wealth and luck. Just wanted to share because I am a massive fan of geology and crystal identification.
-
-
biz.libretexts.org biz.libretexts.org
-
The assumption that women are more relationship oriented, while men are more assertive, is an example of a stereotype.
How’s this stereotype LMAO. It’s just a fact of biological reality
-
-
social-media-ethics-automation.github.io social-media-ethics-automation.github.io
-
While this example is not on social media, I think that something similar is our use of plastic in our everyday lives. On the surface, it's just a bottle of water or a bag of chips, but the reality is that plastic has now permeated into our lives at a microscopic scale.
-
-
social-media-ethics-automation.github.io social-media-ethics-automation.github.io
-
In this screenshot of Twitter, we can see the following information: The account that posted it: User handle is @dog_rates User name is WeRateDogs® User profile picture is a circular photo of a white dog This user has a blue checkmark The date of the tweet: Feb 10, 2020 The text of the tweet: “This is Woods. He’s here to help with the dishes. Specifically, the pre-rinse, where he licks every item he can. 12/10” The photos in the tweet: Three photos of a puppy on a dishwasher The number of replies: 1,533 The number of retweets: 26.2K The number of likes: 197.8K
This gives a brief information of the user's information, the data such as likes and comments gives a brief idea that is this content worth to pay a close reading or not. Clearly this example is worthy! Also just one post it contains soo many information, it's kind of surprising.
-
-
social-media-ethics-automation.github.io social-media-ethics-automation.github.io
-
If we download information about a set of tweets (text, user, time, etc.) to analyze later, we might consider that set of information as the main data, and our metadata might be information about our download process, such as when we collected the tweet information, which search term we used to find it, etc.
I never realized how powerful metadata can be. It’s interesting that it’s not just about the content of the tweets, but also about information like when and how we collected them. That extra layer can really change how we understand and analyze data. It can reveal what time someone does things, trends, and behavior that we don't see behind the scenes.
-
-
social-media-ethics-automation.github.io social-media-ethics-automation.github.io
-
Images are created by defining a grid of dots, called pixels. Each pixel has three numbers that define the color (red, green, and blue), and the grid is created as a list (rows) of lists (columns).
It’s cool to see how images are really just grids of pixels with RGB values, and even something like microRGB fits into that same idea of breaking color down into tiny components. Thinking about images this way makes them feel a lot less mysterious and more like something you can actually work with in code.
-
Sounds are represented as the electric current needed to move a speaker’s diaphragm back and forth over time to make the specific sound waves. The electric current is saved as a number, and those electric current numbers are saved at each time point, so the sound information is saved as a list of numbers.
It’s interesting to think about how sound is really just a list of numbers that tell a speaker how to move, moment by moment, to recreate a noise or a voice. Once you see it that way, audio feels a lot less abstract and more like something you can store, edit, and mess with just like any other data.
-
Images are created by defining a grid of dots, called pixels. Each pixel has three numbers that define the color (red, green, and blue), and the grid is created as a list (rows) of lists (columns).
I've heard only three colors (RGB) are needed because these colors are enough to recreate what the human eye perceives. I think it’s interesting how such a wide range of colors and detailed images can be created just by changing the intensity of three simple values in each pixel.
-
-
social-media-ethics-automation.github.io social-media-ethics-automation.github.io
-
In our example tweet we can see several places where data could be saved in lists:
This section makes it click that a “tweet” isn’t just one thing—it’s basically a bundle of lists (a list of images, a list of likes, a list of replies, etc.). Thinking of it that way also helps explain why social media data gets huge fast, because each post can point to multiple growing lists. It’s kind of wild that even something simple like “who liked this” is literally stored as a list of accounts behind the scenes.
-
-
www.biorxiv.org www.biorxiv.org
-
Note: This preprint has been reviewed by subject experts for Review Commons. Content has not been altered except for formatting.
Learn more at Review Commons
Referee #1
Evidence, reproducibility and clarity
Summary:
This study by Neupane et al. investigates modulators of α-synuclein aggregation, focusing on Ser129-phosphorylated α-synuclein (pSyn129), a pathological hallmark of Parkinson's disease (PD). The authors performed high-content image-based, arrayed CRISPR activation (CRISPRa) and knockout (CRISPRo) screens targeting > 2300 genes related to mitochondrial function, intracellular trafficking, and cytoskeletal reorganization. Using α-Syn overexpressing HEK293 cells, they identified OXR1 and EMC4 as novel modulators of pSyn129 abundance. Key findings were that activation of the mitochondrial protein OXR1 increased pSyn129 by decreasing ATP levels, while ablation of the ER-associated protein EMC4 reduced pSyn129 by enhancing autophagic flux and lysosomal clearance. These findings were validated in human iPSC-derived cortical and dopaminergic neurons.
My major comments have to do with statistical methods and with significance of their findings.
Major comments:
Are the claims and the conclusions supported by the data or do they require additional experiments or analyses to support them?
The claims and conclusions are generally well-supported by the presented data. The dual CRISPRa/CRISPRo screen provides a robust initial discovery platform, and the validation in iPSC-derived neurons strengthens the findings and their translational relevance. The mechanistic insights into OXR1 (ATP levels) and EMC4 (autophagic flux, lysosomal clearance) are supported by the described experiments. The use of two antibodies (81A and EP1536Y) for pSyn129 also enhances confidence in the measurements. I had a few questions about the statistical methods. The main concern I have about methodology for the screen is whether the authors have corrected for multiple hypotheses in their discovery screen. This is not clear from the text, methods, or legends (for Figures 2A/2B/2C).
- Figure 1B suggests a very large range of activation (multiple orders of magnitude) in the initial screen. What is the relationship between level of expression change and functional effect across the screen? How upregulated/downregulated are OXR1 and EMC4 at the mRNA and protein levels?
- Supplemental Figure S2D: Why do the non-targeting controls differ from the majority of the CRISPRa genes? If I am reading the figure correctly, it seems strange that the vast majority of the CRISPRa gene targets reduces pSyn pathology relative to the non-targeting controls (which is why I am wondering whether the level of increased expression correlates with the level of functional effect).
- In Figure 2A/B/C, is the p-value adjusted in any way for multiple comparisons? If so, this should be indicated in the legend. If not, why not? (The potential for false positives in a screen is very large and requires correction for multiple comparisons.)
- Figure 3: It's interesting that different seeding materials have different effects. However, it's quite surprising that the authors find less seeding with MSA-derived material in both the CRISPRa and CRISPRo context. This contradicts the work of Peng and coauthors (PMID 29743672) who find that MSA-derived material is much more potent in seeding aggregates in a number of different cell types. Do the authors have any thoughts about why this is the case?
- Figure 7A: pSyn129 image in the non-targeting control is poor - the very bright dots look like artifact. Not clear why the authors don't corroborate with EP1536Y antibody as they do in Figure 5.
- Overall methodology: Are the pSyn inclusions soluble? This could be easily determined by performing 1% TritonX extraction, for example, and it helps us understand how "pathological" the inclusions are.
- OPTIONAL: The authors perform some interesting experiments looking at genes affected downstream by, for example, OXR1 over-expression. It would be useful to understand whether the upstream effect is dependent on downstream effect. This could be tested by performing double perturbations (e.g. OXR1 overexpression and CCL8 knockout or ALDOC upregulation).
- OPTIONAL: The link between EMC4 ablation and enhanced ER-driven autophagic flux/lysosomal clearance could be corroborated with additional experiments. E.g.: Does EMC4 normally inhibit this pathway? Or only in the context of aSyn fibril seeding?
Are the suggested experiments realistic in terms of time and resources?
The OPTIONAL experiments are generally feasible as they employ methods that the lab is already using in this paper.
Are the experiments adequately replicated and statistical analysis adequate?
See comment about multiple hypothesis testing above.
Significance
This is a well-designed, difficult-to-accomplish study that expands the landscape of pS129Syn modulators. The validation of the primary hits identified in HEK293 cells in iPSC-derived neurons gives the findings greater relevance.
Strengths:
- Novelty: Using an unbiased and high-throughput approach, the study identifies two novel regulators of α-Syn aggregation, namely OXR1 and EMC4.
- Methodological Rigor: The use of arrayed CRISPRa/CRISPRo screens with high-content imaging is powerful and difficult to accomplish. Methodologically, this is a tour de force.
- Orthogonal Validation: The use of multiple α-Syn fibril polymorphs/strains and different antibodies (81A, EP1536Y) strengthens the robustness of the findings.
Limitations:
- It's not clear to me that pSyn129 is the ultimate readout. At a minimum, we should know something about the solubility of the inclusions. Some panels (e.g. Figure 7A) are not very informative in terms of what the authors are calling pSyn129+.
- The study relies on in vitro cellular models. While iPSC-derived neurons are relevant, the complexity of the brain environment, including glial cell interactions is not fully captured. This is fine for an initial report, but it does limit the significance.
- OXR1 and EMC4 seem to be very generic modulators. It's not clear to me that their effects are specific to aSyn or to PD in any way - they might just be effects on very basic cellular functions that would be applicable to a number of stressors or proteinopathies. Maybe that is fine (we probably need to get rid of tau aggregates, too!), but I don't think the authors can claim that they have identified "organelle-specific genetic nodes of aSyn pathology" since they biased their screen towards mitochondria and they don't test any other pathological aggregates. Moreover, from a translational perspective, it's not clear to me that implicating the antioxidant pathway or lysosomal/autophagosomal pathways in the pathogenesis of PD is new, and it's not clear that the specific genes identified would make good therapeutic targets.
-
-
social-media-ethics-automation.github.io social-media-ethics-automation.github.io
-
Examples of Bots
The section on antagonistic bots was especially interesting to me. It’s concerning how bots can create the illusion of mass support or backlash, even when most real users don’t feel that strongly. This makes me think that bots don’t just add noise, but can actually change how people interpret public opinion.
-
We also would like to point out that there are fake bots as well, that is real people pretending their work is the result of a Bot. For example, TikTok user Curt Skelton posted a video claiming that he was actually an AI-generated / deepfake character:
As someone who's majoring in a creative field, I find it both incredibly interesting and concerning just how advanced AI is getting, and where this rapid innovation will take us in just a few years. It's so jarring to be watching a video on Tiktok or Instagram and fully believe it to be completely real, just to feel the need to dissect the video to see if it's really real. I can't begin to imagine how the job industry will change due to AI, but with innovation there (hopefully) comes opportunity.**
-
-
social-media-ethics-automation.github.io social-media-ethics-automation.github.io
-
I think that tech giants are not invested in ethical changes because there is no real risk for them. At the end of the day, the goal is to make more money for shareholders, and if the well-being of users is not going to make them money, then it is not worth investing in. I believe that technology can be regulated at the same speed as its advancement. It's just, regulation will put up barriers to profit, so instead surface-level ethics are presented to us as a bandaid to ethical issues that are growing like a cancer at the same rate technology is advancing.
-
-
Local file Local file
-
In neoclassical and neoliberal economics, state spendingon services is framed as being paid for by the “productive” –i.e. profitable – sector of the economy. But, as we learned inthe pandemic, the most useful and essential parts of our soci-ety are often the least profitable and their workers the leastremunerated. Many profitable sectors are not useful, andoften quite damaging. Large parts of the hyper-profitablefinance sector are parasitic on the public service sector. AsI argued in Chapter 2, this “productive” economy relies on awhole set of disavowed systems – education, domestic labour,environment – without which its profits would be impossible.
I feel this argument, against bullshit jobs, is much stronger than "capitalism has failed"... for once, because it doesn't necessitate a defeatist starting point, and can me framed as even a satirical position (CEOs don't do crap, is laughable), and for second, because just world hypothesis conservatism bias tells people this can't be the case. Messages online tell people "capitalism+democracy" is the way to go, or else dictatorships by the ultra rich (?). I find it amusing, that more of the same sells so nicely, but that's what people see, survivorship bias big companies employing lots of people, and capitalism lifting out pop stars through financial mobility, and the system delivering all the goods we can possible think of. It's become spectacular consumerism, and culture is everywhere. Culture works, it fucking does, but not your culture, rather Gmail's, and Meta, and OnlyFans, Roblox, etc. one.
-
Health, education,housing, transport, food, energy, water, basic communicationsystems – should these not be fixed before we get to culture?1In what follows I make an argument for the centrality ofculture in overcoming the present crisis, and in the followingchapters sketch out what such an agenda might look like. Weneed to acknowledge the “legitimation crisis” to which culture-as- industry was a response – art associated with elitism andpatrician subsidy, the growth of the culture industries
But culture is health! A culture of health, of dancing, of being engaged in mentally challenging projects. Culture is education, it teaches, it creates communities. Culture might not be food per se, but it can help grow food, and cook it, collectively, with shared non-contaminating gardens, and recycling processes, through rites more often depicted in religion.
Culture is communication when misscomunication is rampant: It's not just posting and sharing the life of your children online, it's being aware of its impacts, checking before sending news, and feeling safe enough to talk about suicide and emotions, or to speak up about a close relative who is harassing you, regardless of gender! Is this not a progressivist culture that does not invisibilise these things to private life?
Or rather, arguably, culture could act as these things. Currently, mass consumption culture is at times an inhibating mass-sterelisation device, and at others, a way to propagate neo-colonialist monopolistic ideals who benefit a wealthy few. It also perpetuates the myth of equality and meritocracy, through ads, and its instrumentalist tokenised portrayal of diverse hires and rampant on your face corruption and ideals buying, ultimately makes change within the system almost impossible.
-
Neoliberalism, a distinct phase of global capitalism thatemerged in the early 1980s, is in crisis. The economic, politi-cal, and social solutions it brought to the prior crisis of Fordistsocial democracy in the 1970s no longer work. That muchmight be broadly agreed. It has become ever clearer since the2008– 10 global financial crisis. Stagnant growth, stalled wages,growing inequality, cost- of-living increases, rising indebtedness,increasing precarity of employment, and the erosion of publicservices are the stuff of the nightly news. This is accompaniedby widespread disaffection and disconnection from the politicalprocess, resulting in alternating moments of political efferves-cence (often as anti-politics) and resigned passivity. This in theface of a growing perception of the deepening hold of powerfulelites over the workings of government, in the form of lobby-ing, political finance, corruption, cronyism, the public–private“revolving door”, and a pervasive cynicism towards techno-cratic political expertise
Yes, but there have been some welfare and rights advances! Think women and LGBTIQA+ rights. There is increasing cost because demographics have changed, it's not just automation and IA giving the capitalists excess capital gains. Polarisation and corruption are a byproduct of mass production, of noisy, post-truth production.
-
-
www.bitbybitbook.com www.bitbybitbook.com
-
The “Internet of Things” means that behavior in the physical world will be increasingly captured by digital sensors. In other words, when you think about social research in the digital age you should not just think online, you should think everywhere.
In my Intro to Information class, I recall my professor discussing the substantial impact that many researchers deem AI to have — an impact that is only comparable to that of the introduction of the Internet. It makes me think about how revolutionary AI feels to be in the midst of its development and how I often overlook this scale when thinking about the internet simply because it's all I've known throughout my upbringing. The digitization and automation of many once manual processes is so commonplace now, I can only imagine how much it'll change in the upcoming years
-
-
socialsci.libretexts.org socialsci.libretexts.org
-
All of these problems indicate that older people should be less happy than younger people. If a sociologist did some research and then reported that older people are indeed less happy than younger people, what have we learned?
I only believe something when there is a hypothesis tested many times and the prediction comes true. Many old people get sick and struggle because they did not take care of themselves when they were young and used different things that made their bodies weak. My point is we can’t just keep predicting things. If we keep predicting, it’s just assuming about things we’re not really sure of yet.
-
Sociology can help us understand the social forces that affect our behavior, beliefs, and life chances, but it can only go so far. That limitation conceded, sociological understanding can still go fairly far toward such an understanding, and it can help us comprehend who we are and what we are by helping us first understand the profound yet often subtle influence of our social backgrounds on so many things about us.
When I first got here in America, there were certain things I believed because of my social background. My husband had a therapist, so I joined his therapy. At first, I just listened to other people’s problems and issues, until the therapist asked me about my own feelings about my family. I had a lot of beliefs like ‘my family is great,’ but when he asked me, I felt doomed because I didn’t know what to say and I didn’t even know the definition of feelings. As we kept working, I started to see the reality. I saw the true color, the true image of my past. I realized I have a lot of trauma, but at least now I can face it. I can see it is there, instead of lying to myself that it is not there. I know it’s not perfect and I still have a lot of issues and a lot of scramble in my life, but at least I can work on it now.
-
-
nmoer.pressbooks.pub nmoer.pressbooks.pub
-
When I was an undergraduate at the University of Florida, I didn’t understand that each academic discipline I took courses in to complete the requirements of my degree (history, philosophy, biology, math, political science, sociology, English) was a different discourse community. Each of these academic fields had their own goals, their own genres, their own writing conventions, their own formats for citing sources, and their own expectations for writing style. I thought each of the teachers I encountered in my undergraduate career just had their own personal preferences that all felt pretty random to me. I didn’t understand that each teacher was trying to act as a representative of the discourse community of their field.
Discipline is and always will be part of any professional or academic career, personally I think that if you want to be successful you have to be organized and disciplined. If I feel it's more challenging, it's because it will help me push myself and see what I'm capable of and how I handle situations under pressure.
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the original reviews.
Public Reviews:
Reviewer #1 (Public review):
Summary:
The authors present a novel usage of fluorescence lifetime imaging microscopy (FLIM) to measure NAD(P)H autofluorescence in the Drosophila brain, as a proxy for cellular metabolic/redox states. This new method relies on the fact that both NADH and NADPH are autofluorescent, with a different excitation lifetime depending on whether they are free (indicating glycolysis) or protein-bound (indicating oxidative phosphorylation). The authors successfully use this method in Drosophila to measure changes in metabolic activity across different areas of the fly brain, with a particular focus on the main center for associative memory: the mushroom body.
Strengths:
The authors have made a commendable effort to explain the technical aspects of the method in accessible language. This clarity will benefit both non-experts seeking to understand the methodology and researchers interested in applying FLIM to Drosophila in other contexts.
Weaknesses:
(1) Despite being statistically significant, the learning-induced change in f-free in α/β Kenyon cells is minimal (a decrease from 0.76 to 0.73, with a high variability). The authors should provide justification for why they believe this small effect represents a meaningful shift in neuronal metabolic state.
We agree with the reviewer that the observed f_free shift averaged per individual, while statistically significant, is small. However, to our knowledge, this is the first study to investigate a physiological (i.e., not pharmacologically induced) variation in neuronal metabolism using FLIM. As such, there are no established expectations regarding the amplitude of the effect. In the revised manuscript, we have included an additional experiment involving the knockdown of ALAT in α/β Kenyon cells, which further supports our findings. We have also expanded the discussion to expose two potential reasons why this effect may appear modest.
(2) The lack of experiments examining the effects of long-term memory (after spaced or massed conditioning) seems like a missed opportunity. Such experiments could likely reveal more drastic changes in the metabolic profiles of KCs, as a consequence of memory consolidation processes.
We agree with the reviewer that investigating the effects of long-term memory on metabolism represent a valuable future path of investigation. An intrinsic caveat of autofluorescence measurement, however, is to identify the cellular origin of the observed changes. To this respect, long-term memory formation is not an ideal case study as its essential feature is expected to be a metabolic activation localized to Kenyon cells’ axons in the mushroom body vertical lobes (as shown in Comyn et al., 2024), where many different neuron subtypes send intricate processes. This is why we chose to first focus on middle-term memory, where changes at the level of the cell bodies could be expected from our previous work (Rabah et al., 2022). But our pioneer exploration of the applicability of NAD(P)H FLIM to brain metabolism monitoring in vivo now paves the way to extending it to the effect of other forms of memory.
(3) The discussion is mostly just a summary of the findings. It would be useful if the authors could discuss potential future applications of their method and new research questions that it could help address.
The discussion has been expanded by adding interpretations of the findings and remaining challenges.
Reviewer #2 (Public review):
This manuscript presents a compelling application of NAD(P)H fluorescence lifetime imaging (FLIM) to study metabolic activity in the Drosophila brain. The authors reveal regional differences in oxidative and glycolytic metabolism, with a particular focus on the mushroom body, a key structure involved in associative learning and memory. In particular, they identify metabolic shifts in α/β Kenyon cells following classical conditioning, consistent with their established role in energy-demanding middle- and long-term memories.
These results highlight the potential of label-free FLIM for in-vivo neural circuit studies, providing a powerful complement to genetically encoded sensors. This study is well-conducted and employs rigorous analysis, including careful curve fitting and well-designed controls, to ensure the robustness of its findings. It should serve as a valuable technical reference for researchers interested in using FLIM to study neural metabolism in vivo. Overall, this work represents an important step in the application of FLIM to study the interactions between metabolic processes, neural activity, and cognitive function.
Reviewer #3 (Public review):
This study investigates the characteristics of the autofluorescence signal excited by 740 nm 2-photon excitation, in the range of 420-500 nm, across the Drosophila brain. The fluorescence lifetime (FL) appears bi-exponential, with a short 0.4 ns time constant followed by a longer decay. The lifetime decay and the resulting parameter fits vary across the brain. The resulting maps reveal anatomical landmarks, which simultaneous imaging of genetically encoded fluorescent proteins helps to identify. Past work has shown that the autofluorescence decay time course reflects the balance of the redox enzyme NAD(P)H vs. its protein-bound form. The ratio of free-to-bound NADPH is thought to indicate relative glycolysis vs. oxidative phosphorylation, and thus shifts in the free-to-bound ratio may indicate shifts in metabolic pathways. The basics of this measure have been demonstrated in other organisms, and this study is the first to use the FLIM module of the STELLARIS 8 FALCON microscope from Leica to measure autofluorescence lifetime in the brain of the fly. Methods include registering the brains of different flies to a common template and masking out anatomical regions of interest using fluorescence proteins.
The analysis relies on fitting an FL decay model with two free parameters, f_free and t_bound. F_free is the fraction of the normalized curve contributed by a decaying exponential with a time constant of 0.4 ns, thought to represent the FL of free NADPH or NADH, which apparently cannot be distinguished. T_bound is the time constant of the second exponential, with scalar amplitude = (1-f_free). The T_bound fit is thought to represent the decay time constant of protein-bound NADPH but can differ depending on the protein. The study shows that across the brain, T_bound can range from 0 to >5 ns, whereas f_free can range from 0.5 to 0.9 (Figure 1a). These methods appear to be solid, the full range of fits are reported, including maximum likelihood quality parameters, and can be benchmarks for future studies.
The authors measure the properties of NADPH-related autofluorescence of Kenyon Cells(KCs) of the fly mushroom body. The results from the three main figures are:
(1) Somata and calyx of mushroom bodies have a longer average tau_bound than other regions (Figure 1e);
(2) The f_free fit is higher for the calyx (input synapses) region than for KC somata (Figure 2b);
(3) The average across flies of average f_free fits in alpha/beta KC somata decreases from 0.734 to 0.718. Based on the first two findings, an accurate title would be "Autofluorecense lifetime imaging reveals regional differences in NADPH state in Drosophila mushroom bodies."
The third finding is the basis for the title of the paper and the support for this claim is unconvincing. First, the difference in alpha/beta f_free (p-value of 4.98E-2) is small compared to the measured difference in f_free between somas and calyces. It's smaller even than the difference in average soma f_free across datasets (Figure 2b vs c). The metric is also quite derived; first, the model is fit to each (binned) voxel, then the distribution across voxels is averaged and then averaged across flies. If the voxel distributions of f_free are similar to those shown in Supplementary Figure 2, then the actual f_free fits could range between 0.6-0.8. A more convincing statistical test might be to compare the distributions across voxels between alpha/beta vs alpha'/beta' vs. gamma KCs, perhaps with bootstrapping and including appropriate controls for multiple comparisons.
The difference observed is indeed modest relative to the variability of f_free measurements in other contexts. The fact that the difference observed between the somata region and the calyx is larger is not necessarily surprising. Indeed, these areas have different anatomical compositions that may result in different basal metabolic profiles. This is suggested by Figure 1b which shows that the cortex and neuropile have different metabolic signatures. Differences in average f_free values in the somata region can indeed be observed between naive and conditioned flies. However, all comparisons in the article were performed between groups of flies imaged within the same experimental batches, ensuring that external factors were largely controlled for. This absence of control makes it difficult to extract meaningful information from the comparison between naive and conditioned flies.
We agree with the reviewer that the choice of the metric was indeed not well justified in the first manuscript. In the new manuscript, we have tried to illustrate the reasons for this choice with the example of the comparison of f_free in alpha/beta neurons between unpaired and paired conditioning (Dataset 8). First, the idea of averaging across voxels is supported by the fact that the distributions of decay parameters within a single image are predominantly unimodal. Examples for Dataset 8 are now provided in the new Sup. Figure 14. Second, an interpretable comparison between multiple groups of distributions is, to our knowledge, not straightforward to implement. It is now discussed in Supplementary information. To measure interpretable differences in the shapes of the distributions we computed the first three moments of distributions of f_free for Dataset 8 and compared the values obtained between conditions (see Supplementary information and new Sup. Figure 15). Third, averaging across individuals allows to give each experimental subject the same weight in the comparisons.
I recommend the authors address two concerns. First, what degree of fluctuation in autofluorescence decay can we expect over time, e.g. over circadian cycles? That would be helpful in evaluating the magnitude of changes following conditioning. And second, if the authors think that metabolism shifts to OXPHOS over glycolosis, are there further genetic manipulations they could make? They test LDH knockdown in gamma KCs, why not knock it down in alpha/beta neurons? The prediction might be that if it prevents the shift to OXPHOS, the shift in f_free distribution in alpha/beta KCs would be attenuated. The extensive library of genetic reagents is an advantage of working with flies, but it comes with a higher standard for corroborating claims.
In the present study, we used control groups to account for broad fluctuations induced by external factors such as the circadian cycle. We agree with the reviewer that a detailed characterization of circadian variations in the decay parameters would be valuable for assessing the magnitude of conditioning-induced shifts. We have integrated this relevant suggestion in the Discussion. Conducting such an investigation lies unfortunately beyond the scope and means of the current project.
In line with the suggestion of the reviewer, we have included a new experiment to test the influence of the knockdown of ALAT on the conditioning-induced shift measured in alpha/beta neurons. This choice is motivated in the new manuscript. The obtained result shows that no shift is detected in the mutant flies, in accordance with our hypothesis.
FLIM as a method is not yet widely prevalent in fly neuroscience, but recent demonstrations of its potential are likely to increase its use. Future efforts will benefit from the description of the properties of the autofluorescence signal to evaluate how autofluorescence may impact measures of FL of genetically engineered indicators.
Recommendations for the authors
Reviewer #1 (Recommendations for the authors):
(1) Y axes in Figures 1e, 2c, 3b,c are misleading. They must start at 0.
Although we agree that making the Y axes start at 0 is preferable, in our case it makes it difficult to observe the dispersion of the data at the same time (your next suggestion). To make it clearer to the reader that the axes do not start at 0, a broken Y-axis is now displayed in every concerned figure.
(2) These same plots should have individual data points represented, for increased clarity and transparency.
Individual data points were added on all boxplots.
Reviewer #2 (Recommendations for the authors):
I am evaluating this paper as a fly neuroscientist with experience in neurophysiology, including calcium imaging. I have little experience with FLIM but anticipate its use growing as more microscopes and killer apps are developed. From this perspective, I value the opportunity to dig into FLIM and try to understand this autofluorescence signal. I think the effort to show each piece of the analysis pipeline is valuable. The figures are quite beautiful and easy to follow. My main suggestion is to consider moving some of the supplemental data to the main figures. eLife allows unlimited figures, moving key pieces of the pipeline to the main figures would make for smoother reading and emphasize the technical care taken in this study.
We thank the reviewer for their feedback. Following their advice we have moved panels from the supplementary figures to the main text (see new Figure 2).
Unfortunately, the scientific questions and biological data do not rise to the typical standard in the field to support the claims in the title, "In vivo autofluorescence lifetime imaging of the Drosophila brain captures metabolic shifts associated with memory formation". The authors also clearly state what the next steps are: "hypothesis-driven approaches that rely on metabolite-specific sensors" (Intro). The advantage of fly neuroscience is the extensive library of genetic reagents that enable perturbations. The key manipulation in this study is the electric shock conditioning paradigm that subtly shifts the distribution of a parameter fit to an exponential decay in the somas of alpha/beta KCs vs others. This feels like an initial finding that deserves follow-up; but is it a large enough result to motivate a future student to pick this project up? The larger effect appears to be the gradients in f_free across KCs overall (Figure 2b). How does this change with conditioning?
We acknowledge that the observed metabolic shift is modest relative to the variability of f_free and agree that additional corroborating experiments would further strengthen this result. Nevertheless, we believe it remains a valid and valuable finding that will be of interest to researchers in the field. The reviewer is right in pointing out that the gradient across KCs is higher in magnitude, however, the fact that this technique can also report experience-dependent changes, in addition to innate heterogeneities across different cell types, is a major incentive for people who could be interested in applying NAD(P)H FLIM in the future. For this reason, we consider it appropriate to retain mention of the memory-induced shift in the title, while making it less assertive and adding a reference to the structural heterogeneities of f_free revealed in the study. We have also rephrased the abstract to adopt a more cautious tone and expanded the discussion to clarify why a low-magnitude shift in f_free can still carry biological significance in this context. Finally, we have added the results of a new set of data involving the knockdown of ALAT in Kenyon cells, to further support the relevance of our observation relative to memory formation, despite its small magnitude. We believe that these elements together form a good basis for future investigations and that the manuscript merits publication in its present form.
Together, I would recommend reshaping the paper as a methods paper that asks the question, what are the spatial properties of NADPH FL across the brain? The importance of this question is clear in the context of other work on energy metabolism in the MBs. 2P FLIM will likely always have to account for autofluorescence, so this will be of interest. The careful technical work that is the strength of the manuscript could be featured, and whether conditioning shifts f_free could be a curio that might entice future work.
By transferring panels of the supplementary figures to the main text (see new Figure 2) as suggested by Reviewer 2, we have reinforced the methodological part of the manuscript. For the reasons explained above, we however still mention the ‘biological’ findings in the title and abstract.
Minor recommendations on science:
Figure 2C. Plotting either individual data points or distributions would be more convincing.
Individual data points were added on all boxplots.
There are a few mentions of glia. What are the authors' expectations for metabolic pathways in glia vs. neurons? Are glia expected to use one more than the other? The work by Rabah suggests it should be different and perhaps complementary to neurons. Can a glial marker be used in addition to KC markers? This seems crucial to being able to distinguish metabolic changes in KC somata from those in glia.
Drosophila cortex glia are thought to play a similar role as astrocytes in vertebrates (see Introduction). In that perspective, we expect cortex glia to display a higher level of glycolysis than neurons. The work by Rabah et al. is coherent with this hypothesis. Reviewer 2 is right in pointing out that using a glial marker would be interesting. However, current technical limitations make such experiments challenging. These limitations are now exposed in the discussion.
The question of whether KC somata positions are stereotyped can probably be answered in other ways as well. For example, the KCs are in the FAFB connectomic data set and the hemibrain. How do the somata positions compare?
The reviewer’s suggestion is indeed interesting. However, the FAFB and hemibrain connectomic datasets are based on only two individual flies, which probably limits their suitability for assessing the stereotypy of KC subtype distributions. In addition, aligning our data with the FAFB dataset would represent substantial additional work.
The free parameter tau_bound is mysterious if it can be influenced by the identity of the protein. Are there candidate NADPH binding partners that have a spatial distribution in confocal images that could explain the difference between somas and calyx?
There are indeed dozens of NADH- or NADPH-binding proteins. For this reason, in all studies implementing exponential fitting of metabolic FLIM data, tau_bound is considered a complex combination of the contributions from many different proteins. In addition, one should keep in mind that the number of cell types contributing to the autofluorescence signal in the mushroom body calyx (Kenyon cells, astrocyte-like and ensheathing glia, APL neurons, olfactory projection neurons, dopamine neurons) is much higher than in the somas (only Kenyon cells and cortex glia). This could also participate in the observed difference. Hence, focusing on intracellular heterogeneities of potential NAD(P)H binding partners seems premature at that stage.
The phrase "noticeable but not statistically significant" is misleading.
We agree with the reviewer and have removed “noticeable but” from the sentence in the new version of the manuscript.
Minor recommendations on presentation:
The Introduction can be streamlined.
We agree that some parts of the Introduction can seem a bit long for experts of a particular field. However, we think that this level of detail makes the article easily accessible for neuroscientists working on Drosophila and other animal models but not necessarily with FLIM, as well as for experts in energy metabolism that may be familiar with FLIM but not with Drosophila neuroscience.
-
-
-
Reviewer #3 (Public review):
This paper applies a computational model to behavior in a probabilistic operant reward learning task (a 3-armed bandit) to uncover differences between individuals with temporomandibular disorder (TMD) compared with healthy controls. Integrating computational principles and models into pain research is an important direction, and the findings here suggest that TMD is associated with subtle changes in how uncertainty is represented over time as individuals learn to make choices that maximize reward. There are a number of strengths, including the comparison of a volatile Kalman filter (vKF) model to some standard base models (Rescorla Wagner with 1 or 2 learning rates) and parameter recovery analyses suggesting that the combination of task and vKF model may be able to capture some properties of learning and decision-making under uncertainty that may be altered in those suffering from chronic pain-related conditions.
I've focused my comments in four areas: (1) Questions about the patient population, (2) Questions about what the findings here mean in terms of underlying cognitive/motivational processes, (3) Questions about the broader implications for understanding individuals with TMD and other chronic pain-related disorders, and (4) Technical questions about the models and results.
(1) Patient population
This is a computational modelling study, so it is light on characterization of the population, but the patient characteristics could matter. The paper suggests they were hospitalized, but this is not a condition that requires hospitalization per se. It would be helpful to connect and compare the patient characteristics with large-scale studies of TMD, such as the OPPERA study led by Maixner, Fillingim, and Slade.
(2) What cognitive/motivational processes are altered in TMD
The study finds a pattern of alterations in TMD patients that seems clear in Figure 2. Healthy controls (HC) start the task with high estimates of volatility, uncertainty, and learning rate, which drop over the course of the task session. This is consistent with a learner that is initially uncertain about the structure of the environment (i.e., which options are rewarded and how the contingencies change over time) but learns that there is a fixed or slowly changing mean and stationary variance. The TMD patients start off with much lower volatility, uncertainty, and learning rate - which are actually all near 0 - and they remain stable over the course of learning. This is consistent with a learner who believes they know the structure of the environment and ignores new information.
What is surprising is that this pattern of changes over time was found in spite of null group differences in a number of aspects of performance: (1) stay rate, (2) switch rate, (3) win-stay/lose-switch behaviors, (4) overall performance (corrected for chance level), (5) response times, (6) autocorrelation, (7) correlations between participants' choice probability and each option's average reward rate, (7) choice consistency (though how operationalized is not described?), (8) win-stay-lose-shift patterns over time. I'm curious about how the patterns in Figure 2 would emerge if standard aspects of performance are essentially similar across groups (though the study cannot provide evidence in favor of the null). It will be important to replicate these patterns in larger, independent samples with preregistered analyses.
The authors believe that this pattern of findings reveals that TMD patients "maintain a chronically heightened sensitivity to environmental changes" and relate the findings to predictive processing, a hallmark of which (in its simplest form) is precision-weighted updating of priors. They also state that the findings are not related to reduced overall attentiveness or failure to understand the task, but describe them as deficits or impairments in calibrating uncertainty.
The pattern of differences could, in fact, result from differences in prior beliefs, conceptualization of the task, or learning. Unpacking these will be important steps for future work, along with direct measures of priors, cognitive processes during learning, and precision-weighted updating.
(3) Implications for understanding chronic pain
If the findings and conclusions of the paper are correct, individuals with TMD and perhaps other pain-related disorders may have fundamental alterations in the ways in which they make decisions about even simple monetary rewards. The broader questions for the field concern (1) how generalizable such alterations are across tasks, (2) how generalizable they are across patient groups and, conversely, how specific they are to TMD or chronic pain, (3) whether they are the result of neurological dysfunction, as opposed to (e.g.) adaptive strategies or assumptions about the environment/task structure.
It will be important to understand which features of patients' and/or controls' cognition are driving the changes. For example, could the performance differences observed here be attributable to a reduced or altered understanding of the task instructions, more uncertainty about the rules of the game, different assumptions about environments (i.e., that they are more volatile/uncertain or less so), or reduced attention or interest in optimizing performance? Are the controls OVERconfident in their understanding of the environment?
This set of questions will not be easy to answer and will be the work of many groups for many years to come. It is a judgment call how far any one paper must go to address them, but my view is that it is a collaborative effort. Start with a finding, replicate it across labs, take the replicable phenomena and work to unpack the underlying questions. The field must determine whether it is this particular task with this model that produces case-control differences (and why), or whether the findings generalize broadly. Would we see the same findings for monetary losses, sounds, and social rewards? Tasks with painful stimuli instead of rewards?
Another set of questions concerns the space of computational models tested, and whether their parameters are identifiable. An alteration in estimated volatility or learning rate, for example, can come from multiple sources. In one model, it might appear as a learning rate change and in another as a confirmation bias. It would be interesting in this regard to compare the "mechanisms" (parameters) of other models used in pain neuroscience, e.g., models by Seymour, Mancini, Jepma, Petzschner, Smith, Chen, and others (just to name a few).
One immediate next step here could be to formally compare the performance of both patients and controls to normatively optimal models of performance (e.g., Bayes optimal models under different assumptions). This could also help us understand whether the differences in patients reflect deficits and what further experiments we would need to pin that down.<br /> In addition, the volatility parameter in the computational model correlated with apathy. This is interesting. Is there a way to distinguish apathy as a particular clinical characteristic and feature of TMD from apathy in the sense of general disinterest in optimal performance that may characterize many groups?
If we know this, what actionable steps does it lead us to take? Could we take steps to reduce apathy and thus help TMD patients better calibrate to environmental uncertainty in their lives? Or take steps to recalibrate uncertainty (i.e., increase uncertainty adaptation), with benefits on apathy? A hallmark of a finding that the field can build off of is the questions it raises.
(4) Technical questions about the models and results
Clarification of some technical points would help interpret the paper and findings further:
(a) Was the reward probability truly random? Was the random walk different for each person, or constrained?
(b) When were self-report measures administered, and how?
(c) Pain assessments: What types of pain? Was a body map assessed? Widespreadness? Pain at the time of the test, or pain in general?
(d) Parameter recovery: As you point out, r = 0.47 seems very low for recovery of the true quantity, but this depends on noise levels and on how the parameter space is sampled. Is this noise-free recovery, and is it robust to noise? Are the examples of true parameters drawn from the space of participants, or do they otherwise systematically sample the space of true parameters?
(e) What are the covariances across parameter estimates and resultant confusability of parameter estimates (e.g., confusion matrix)?
(f) It would be helpful to have a direct statistical comparison of controls and TMD on model parameter estimates.
(g) Null statistical findings on differences in correlations should not be interpreted as a lack of a true effect. Bayes Factors could help, but an analysis of them will show that hundreds of people are needed before it is possible to say there are no differences with reasonable certainty. Some journals enforce rules around the kinds of language used to describe null statistical findings, and I think it would be helpful to adopt them more broadly.
(h) What is normatively optimal in this task? Are TMD patients less so, or not? The paper states "aberrant precision (uncertainty) weighting and misestimation of environmental volatility". But: are they misestimates?
(i) It's not clear how well the choice of prior variance for all parameters (6.25) is informed by previous research, as sensible values may be task- and context-dependent. Are the main findings robust to how priors are specified in the HBI model?
-
-
www.scientificamerican.com www.scientificamerican.com
-
The real world consequences are stacking up: the trans military ban, bathroom bills, and removal of workplace and medical discrimination protections, a 41-51 percent suicide attempt rate and targeted fatal violence . It’s not just internet trolling anymore.
The author's purpose in writing this article is clearly laid out in the opening portion of this document. We can see that her main motivation is to uplift that trans community while educating the masses on the trans struggle due to the everlasting backlash that the trans community gets.
-
The irony in all this is that these “protectors of enlightenment” are guilty of the very behavior this phrase derides. Though often dismissed as just a fringe internet movement, they espouse unscientific claims that have infected our politics and culture. Especially alarming is that these “intellectual” assertions are used by nonscientists to claim a scientific basis for the dehumanization of trans people. The real world consequences are stacking up: the trans military ban, bathroom bills, and removal of workplace and medical discrimination protections, a 41-51 percent suicide attempt rate and targeted fatal violence . It’s not just internet trolling anymore.Contrary to popular belief, scientific research helps us better understand the unique and real transgender experience. Specifically, through three subjects: (1) genetics, (2) neurobiology and (3) endocrinology. So, hold onto your parts, whatever they may be. It’s time for “the talk.”
This text is about the use of scientific research to understand the unique and real trans experience using genetics, neurobiology, and endocrinology. Its main message is that the trans experience is far more complicated than many dehumanizing articles make it out to be.
-
It’s not just internet trolling anymore.
The text cites ongoing U.S. debates over transgender rights, showing how scientific arguments are being used to justify legislation and social exclusion. While also showing the broader social and political context that online rhetoric has real-world consequences.
-
Let’s just take the most famous example of sexual dimorphism in the brain: the sexually dimorphic nucleus of the preoptic area (sdnPOA). This tiny brain area with a disproportionately sized name is slightly larger in males than in females. But it’s unclear if that size difference indicates distinctly wired sdnPOAs in males versus females, or if—as with the bipotential primordium—the same wiring is functionally weighted toward opposite ends of a spectrum. Throw in the observation that the sdnPOA in gay men is closer to that of straight females than straight males, and the idea of “the male brain” falls apart.
This small paragraph displays that the text is meant for college students and also individuals familiar with biology, and can understand scientific reasoning from research. We also know because most likely, those who are subscribing to the SCI AM are generally interested in or have a background in STEM/science
-
The real world consequences are stacking up: the trans military ban, bathroom bills, and removal of workplace and medical discrimination protections, a 41-51 percent suicide attempt rate and targeted fatal violence . It’s not just internet trolling anymore.
Exigence: The real world changes in policies and increase of hate crime was a primary motivator for the author writing this text. They highlight that this has been a longstanding issue with specifically the scientific community critiquing the existence of being transgender, though the "real world consequences" drove D Sun to write about this issue.
-
-
www.biorxiv.org www.biorxiv.org
-
Reviewer #2 (Public review):
Summary:
The goal of the experiment was to identify the fMRI neural correlates of persistence and recovery of forgotten memories. A forgotten memory was defined behaviorally as successful learning, followed by failure in a recall format task, followed by next-day success in a recognition format task. The comparison is to memories that were not forgotten at any stage of the task. Various univariate, connectivity, and multivariate analyses were used to identify neural correlates of forgotten memories that were recovered, that remained forgotten, and successful memory. Some claims are made about how activity of the "episodic memory network" predicts the persistence of forgotten memories.
Strengths:
Studies on the persistence of forgotten memories in rodent models have been used to make some novel claims about the potential properties of engrams. Attempting similar research in humans is a laudable goal.
Patterns of behavioral responses are consistent across subjects.
Weaknesses:
I do not find that the fMRI results fit the narrative provided.
A major issue is that primary results do not replicate across the two fMRI datasets that were collected using the same task. For example, hippocampal activity associated with correct responses (confident and guess) was identified in the group receiving the fMRI scan that used a small FOV, but not in the group that received an fMRI scan of the whole brain, for both 30-min and 24-hr delays (lines 202-217). This suggests that the main findings are not even replicable internally within the same experiment. There is no reasonable justification for this.
Next, most of the reported fMRI findings do not meet reasonable thresholds for statistical significance. In many places, the authors acknowledge this in the text by saying that a difference in the fMRI metric "tended towards significant correlation" or that comparisons "revealed non-significant mean value comparisons". It is not clear why these non-significant findings are interpreted as though they are positive findings. Beyond that, many of the reported findings are not meeting the threshold (i.e., p=0.058), without any acknowledgement that they are marginal. Beyond that, the majority of comparisons that are interpreted in the main text are not significant based on the companion information provided in the supplementary tables. That is, they are totally non-significant when using FWE or FDR correction at either the cluster or peak levels.
Beyond this, the supplementary tables indicate that "clusters identified solely within white matter regions have been excluded." The fact that there are any findings in white matter to ignore indicates that the statistical thresholds are inappropriate. It's tantamount to seeing activation in the brain of a dead fish.
The overall picture based on these factors is that the statistical tests did not use sufficiently stringent safeguards against false positives given the multiple comparison problem that plagues fMRI. So, there are tons of false positives, which are being selectively interpreted to tell a particular story. That is, each comparison yields lots of findings in many brain area, and those that do not fit the particular narrative are being ignored (including those in white matter). What's more, when the small FOV fMRI scan is done, the imaging volume is centered on the hippocampus and its close network, so all false positives appear to be exactly in those brain regions about which the authors want to make conclusions. When throwing darts, you will always hit a bullseye if that is all that exists. The fact that the same comparisons done in the companion whole-brain dataset do not yield the same results is telling: the analysis plan is not sufficiently rigorous to yield findings that are replicable.
Further, I think that it is highly debatable whether the task measures the recovery of forgotten memories at all. Forgotten memories are defined as those that fail when tested using a recollection format but succeed when tested using a recognition format. The well-characterized distinction between recollection and recognition is thus being construed as telling us something about the fate of engrams. I think the much more likely alternative is that "forgotten" memories are just relatively weak memories that don't meet whatever criteria subjects typically use when making recollection judgments, and not some special category of memory. In terms of brain activation, they seem for the most part to follow the pattern of stronger memory, but weaker.
Finally, many hypotheses are used as though they are proven. For instance, fMRI activity patterns are called "engrams" even though there are no tests to determine whether they meet reasonable criteria that have been adopted in the engram literature (e.g., necessity, sufficiency). Whatever happens over the 24-hour delay is called "consolidation" even if there is no test that consolidation has occurred. Etc. It becomes hard to differentiate what is an assumption, versus a hypothesis, versus an inference/conclusion.
-
-
dbmi7byi6naiyr.archive.is dbmi7byi6naiyr.archive.is
-
“It’s not that different than looking at the printing press, and the evolution of the book,” he said. After Gutenberg, the printing press was mostly used to mimic the calligraphy in bibles. It took nearly 100 years of technical and conceptual improvements to invent the modern book. “There was this entire period where they had the new technology of printing, but they were just using it to emulate the old media.”
Tags
Annotators
URL
-
-
-
On Typewriters: Condition is King; Context is Queen.
It bears mentioning that an expert/professional repair person can only tell very little of the condition of a typewriter by photos. Does it look generally clean? Are the decals in tact? Does the segment look clean (a vague proxy for the potential condition of the internals)? Is anything obvious missing (knobs, return lever, keys)? Does it look cared for or has it been neglected in a barn for half a century? Most modern typewriters made after 1930 in unknown condition are worth about $5-25 and they peak at about $500 when purchased from a solid repair shop unless some Herculean additional restoration has taken place, they've got a rarer typeface, or are inherently actually rare. Hint: unless it's a pro repair shop or very high end collector with lots of experience, don't trust anyone saying that a typewriter is "rare", run the other direction. Run faster if they say it "works, but just needs a new ribbon" as—even at the most expensive—new ribbon is only $15 and their "rare" $600+ machine should have fresh, wet ribbon. The rule of thumb I use is that no one online selling a typewriter knows anything about it, including if it actually works. Worse, they've probably priced it at professional repair shop prices because they don't know that condition is king.
The least experienced typist will know far more about the condition of a machine by putting their hands on it and trying it out. Does it generally work? Does the carriage move the full length of its travel? Can you set the margins at the extremes and space reliably from one end to another? Does it skip? Is the inside clean or full of decades of dried oil, dust, and eraser crumbs? Does the margin release work? Does it backspace properly? If typing HHHhhhHHH are the letters all printed well and on the same baseline?
Presumably a typewriter at an antique store will meet these minimum conditions (though be aware that many don't as their proprietors have no idea about typewriters other than that if they wait long enough, some sucker will spend $150 on almost anything). They've done the work of finding a machine that (barely) works, housing it, and presenting it to the public for sale. This time and effort is worth something to the beginning typewriter enthusiast, but worth much, much less to the longer term practiced collector.
If everything is present and at least generally limping along, you've got yourself a $30 typewriter. Most people can spend a few hours watching YouTube videos and then manage to clean and lubricate a typewriter to get it functioning reasonably. You can always learn to do the adjustments from Youtube videos. (Or just take it to a repair shop and fork over $200-400 to get things squared.)
If you're getting into collecting, you'll make some useful mistakes by overpaying in the beginning and those mistakes will teach you a lot.
Maybe you're a tinkerer and looking for a project? If so, then find the cheapest machine you can get your hands on (maybe a Royal KMM for $9 at thrift) and work your way through a home study course.
Otherwise, if you're just buying one or two machines to use—by far—the best value you'll find is to purchase a cleaned, oiled, and well-adjusted machine from a repair shop. Sure it might cost $350-600, but what you'll save in time, effort, heartache, repair, etc. will more than outweigh the difference. Additionally you'll have a range of machines to choose from aesthetically and you can test out their feel to find something that works best for you.
Or, you could buy a reasonable machine like this for $70 and find out it needs cleaning, oiling, and adjusting and potentially a few repairs. The repair tab might run you an additional $450. Is it worth it when a repair shop would have sold you the same or a very similar machine in excellent condition for $350?
Remember in asking about the cost and value of a typewriter, you're actually attempting to maximize a wide variety of variables including, but not limited to: upfront money, information about the current state of the market, information/knowledge about the machine itself, information about how to clean it, information about oiling it, information about adjusting, information about repairing it, cost and availability of tools and repair parts, and the time involved for both learning and doing all of these. The more time you've spent learning and doing all of these, the better "deals" you'll find, but gaining this expertise is going to cost you a few years of life. What is all this "worth" when you just want to type on a machine that actually works?
Most of the prognostication you'll find in fora like this will be generally useless to you because you're not readily aware of the context and background of the respondents with respect to all of the variables above. Similarly they're working with no context about you, your situation, where you live, what's available in your area, your level of typewriter knowledge, or your budget. You don't know what you don't know. At the end of the day, you're assuredly just as well off to use a bit of your intuition and putting your hands on a machine and trying it out. Then ask: "What is it worth to you?"
If you're simply asking: "Is this highway robbery?", the answer is no.
More resources (and some of my own context) if you need them: https://boffosocko.com/research/typewriter-collection/
Happy typing.
Reply to u/NeverTheNess at https://reddit.com/r/typewriters/comments/1q7eho6/spotted_a_royal_at_an_antique_store_good/<br /> RE: run-of-the-mill late 70s plastic Litton/Royal typewriter
-
-
www.biorxiv.org www.biorxiv.org
-
Reviewer #1 (Public review):
Summary:
This study was designed to manipulate and analyze the effects of chemosensory cues on visuomotor control. They approach this by analyzing how eye-body coordination and brain-wide activity are altered with specific chemosensation in larval zebrafish. After analyzing the dynamics of coupled saccade-tail coordination sequences - directionally linked and typically coupled to body turns - the authors investigated the effects of sensory cues shown to be either aversive or appetitive on freely swimming zebrafish on the eye-body coordination. Aversive chemicals lead to an increase in saccade-tail sequences in both number and dynamics, seemingly facilitating behaviors like escape. Brain-wide imaging led the authors to neurons in the telencephalic pallium as a target to study eye-body coordination. Pallium neuron activity correlated with both aversive chemicals and coupled saccade-tail movements.
Recommendations for improvement are minimal. So much of the data is ultimately tabular, and the figures are an impenetrable wall of datapoints. 1c is an excellent example: three concentrations are presented, but it's rare for the three averages to trend appropriately. The key point, which is that aversive odors are repulsive and attractive odors (sometimes) attractive just gets lost in showing the three concentrations individually; it also makes direct comparisons impossible. There are similar challenges abound in the violin plots in 4e-4h, the error bars on the "fits" in 4i-4m, and so on. We recommend selecting an illustrative subset of data to present to permit interpretation and putting the rest in a supplemental table. (Presenting) less is more (effective).
-
-
www.medrxiv.org www.medrxiv.org
-
R0:
Reviewer #1:
I reviewed the manuscript titled ‘Awareness of anthrax disease and the knowledge of its transmission and symptoms identification: a cross sectional study among butchers in Ile-Ife’ and below are my observations: Abstract The authors were not consistent in the use of tenses. They should stick to the use of past tenses or continuous tenses. In line 43 they mentioned ‘were not aware’ but in 45 used ‘are aware’. Aware ‘of’ is more preferable than aware ‘about’. They were also not consistent in the use of singular or plural words. For instance, in line 46, they referred to ‘animal’ and then ‘animals’. They can stick to the former as a collective noun or the latter as plural. The authors’ conclusion that there was poor awareness is not consistent with the fact that there was no mention of scoring the questionnaire in the methods. In addition, the lack of mention of knowledge which is a part of the title in the abstract leaves a major void. Knowledge should be captured both in the methods and results. Otherwise, it should be deleted from the title. Introduction Remove the space between the punctuation and the citation in square bracket see lines 63, 79 and 112, for instance. Anthrax is a disease (not an agent) caused by Bacillus anthracis not ‘Bacillus Anthrax’. Line 75: Change ‘buy’ with ‘but’. Authors should stick to ‘human or ‘humans’ (Lines 74 and 76). The transition from one paragraph to the other is not smooth. For instance, lines 79 and 80 do not have any connection. In fact paragraph 4 has no place in its current position in the background of the study. The first sentence of paragraph 5 makes no sense and should be recast. The statement that the first case of anthrax in Nigeria was in 2023 is a lie! The authors should make use of the correct words: For instance, ‘body openings’ should be referred to as ‘body orifices’. Line 104: The authors seem to have changed from reporting a study to writing a proposal. Lines 105 – 108 require referencing. Lines 111 and 112: Change ‘has’ to ‘have’. Line 118: ‘Osun - State, South-Western’, should be written ‘Osun State, southwestern’. Line 121: Change ‘it's’ to ‘its’. Line 125: Change ‘accommodates’ to ‘accommodate’. The authors should draw a map of the area being described. Line 134: Change ‘accommodates’ to ‘accommodate’. The inclusion and exclusion criteria are confusing: ‘Butchers who were standing were included’. Does this mean that those butchers who were sitting were excluded? ‘Butchers who were sick were excluded’, therefore only the healthy were included. How did the authors identify healthy and sick butchers? What was the reason for the chosen criteria? Lines 140 - 149: The fact that the authors are contemplating calculating sample size and recruiting respondents for a study that is concluded is absurd. Lines 158 and 159: Change ‘was’ to ‘were’. Line 161: Which random sampling did the authors use to select the butchers? They should be specific in explaining how the sampling method used was applied in the work. The sampling was not well explained. The authors claimed to have used multi-stage sampling technique. What was the sampling frame in each stage and how were they prepared or accessed? The authors did not explain whether and how the questionnaire was validity and reliability tested. Was there a pilot study and how was it conducted? Without these, the instrument is not valid for a meaningful research. How many sections did the instrument contain and how many questions were there in each section? Was the questionnaire scored and how was it scored? Without scoring the questionnaire, the authors cannot determine the levels of awareness or knowledge. Results Line 167: Change ‘questionnaires’ to ‘questionnaire’ because only one questionnaire was used for the study. Line 174: Change ‘rare’ to ‘rear’. The authors claimed to have worked with only commercial butchers. How come most of them have become cattle rearers? Table 1: Change ‘live stock’ to ‘livestock’. Line 180 – 181. Recast the sentence. Discussion The authors seem not to understand that there is a difference between knowledge and awareness, Therefore, it may be worthwhile to define both words in the work. The authors’ claims on knowledge level may not be correct without scoring knowledge to determine the level among the participants. Comparisons made with some author, example Cadmus et al. (2024) do not rhyme.
Limitations Secondary education is not low level of literacy. Therefore, an area that has more than 70% of the population that attained secondary education cannot be regarded as having low level of literacy.
Reviewer #2:
The manuscript “Awareness of Anthrax Disease and the Knowledge of Its Transmission and Symptoms Identification: A Cross-Sectional Study Among Butchers in Ile-Ife” addresses an important public health issue, the awareness and knowledge of anthrax among butchers, that represents a high-risk occupational category in Nigeria. The topic is relevant, timely, also including a One Health perspective. The study provides useful descriptive data that could inform public health education and prevention strategies. The authors present clear findings about the low awareness of anthrax among butchers and the implications for public health. The study is quite good in methodology and analysis, but there are different areas for improvement, before the manuscript could be considered for publication. As regards the study design, it would be appropriate to clarify the sampling frame, randomization process, and justify the adequacy of the final sample size. How were markets and butchers selected? Was any sampling frame used? No basic inferential analyses (e.g., chi-square tests) were performed to identify factors associated with awareness and knowledge. Could you please, add this kind of analyses? Despite collecting rich demographic data and having explicit research objectives to assess awareness, no inferential analyses examine whether education, income, or animal sourcing practices associate with knowledge levels. Chi-square tests and logistic regression would identify high-risk subgroups requiring targeted interventions, fulfilling the study's stated aims and enhancing public health utility beyond simple prevalence reporting The results could be enriched by analysing possible relationships between the respondents' demographics (e.g., education, age) and their awareness or knowledge of anthrax. This would help contextualize the findings. The discussion could be strengthened by addressing any potential biases in the study (e.g., recall bias due to the use of self-reported data). The paper could explore in greater depth why the awareness was low (e.g., cultural factors, lack of effective public health messaging). While the conclusion addresses the need for improved awareness, it could be more specific about what measures should be taken. For example, the authors could suggest specific communication strategies, community-based interventions, or policy changes. The figures could benefit from more detailed captions that explain what each figure illustrates beyond just showing the data. Furthermore, the manuscript contains grammatical errors and spelling mistakes throughout such as “symtoms instead of symptoms in the short title” “Bacillus Anthrax (line 67)” instead of Bacillus anthracis, “unclothed blood (line 83)” instead of “unclotted blood”. Unclotted is better than un-clotted. Line 217 4got? Correct. Write B. anthracis in italics all over the manuscript. Some sentences are unclear or repetitive, especially in the Background and Discussion sections, so a professional English language editing is recommended.
Academic Editor:
The reviewers have raised a number of concerns that need attention. In particular, they request additional information on methodological aspects of the study, additional analysis, a clearly defined research question, and improvements to the quality of the language used.
-
-
social-media-ethics-automation.github.io social-media-ethics-automation.github.io
-
Something is right or wrong because God(s) said so. Euthyphro Dilemma: “Is the pious [action] loved by the gods because it is pious, or is it pious because it is loved by the gods?” (Socrates, 400s BCE Greece) If the gods love an action because it is morally good, then it is good because it follows some other ethics framework. If we can figure out which ethics framework the gods are using, then we can just apply that one ourselves without the gods. If, on the other hand, an action is morally good because it is loved by the gods, then it doesn’t matter whether it makes sense under any ethics framework, and it is pointless to use ethics frameworks.1
As someone who grew up in a religious household, I often asked questions challenging this theory. It's interesting to think about what could be reprimanded or praised by your god(s)/religious circle, as long as it was written into the guidelines in a scripture or reading. Additionally, I do think that this thinking is dangerous, as it opens up the possibility for people within the religion to misinterpret or maliciously translate certain texts to push negative propaganda to a group of people, and the possibility of mistranslation is incredibly high as most of these texts were written hundreds of years ago.
-
-
social-media-ethics-automation.github.io social-media-ethics-automation.github.io
-
We also see this phrase used to say that things seen on social media are not authentic, but are manipulated, such as people only posting their good news and not bad news, or people using photo manipulation software to change how they look
I think this is an interesting concept to think about, as we are usually conditioned to think that the internet "isn't real", that most things online are fabricated, exaggerated, etc. However, I do think that just because this is common online, it's not to say that "real life" is a place where everyone is completely authentic and themselves, as some people may feel that they only want to share the good parts of their lives with their friends or family, while keeping anything that wouldn't be considered "good" to themselves, and vice versa. I do think it's hasty to say that all that we see on social media "is not real", as there are plenty of real people behind each account, but we must consider that because people are able to be behind potentially anonymous accounts, it is much easier to fabricate stories or life experiences, or to center one's entire online presence around a portion of their life they want the internet to see, essentially artificially creating an online persona that is not reflective of who they are in real life.
-
-
unpublishablepapers.substack.com unpublishablepapers.substack.com
-
Meanwhile, 31% of 8-12 year olds have spoken with large language models. 23% have talked to strangers online, while only 44% have physically spoken to a neighbor without their parents. 50% have seen pornography by the time they turn 13.
Kids are independent in the digital world before the physical one.
That flips the historical order. Earlier generations learned:
- physical world → social norms → abstract/digital spaces
Now it’s:
- abstract/digital spaces → simulated interaction → limited real-world agency
This matters because:
**Digital spaces forgive mistakes cheaply
Physical spaces teach consequences viscerally** - “Physical spaces teach consequences viscerally”
In physical space, mistakes hit the nervous system, not just the mind.
Run too fast → you fall. Ignore a curb → you trip. Misjudge a car → you don’t get a second try. Shout too loud → heads turn. Cross a boundary → someone reacts in front of you.
The feedback is:
Immediate
Embodied
Often irreversible
Digital spaces teach what you can get away with. Physical spaces teach what actually happens.
Both are necessary. But they are not interchangeable.
-
-
socialsci.libretexts.org socialsci.libretexts.org
-
Why not just listen to a recording of the lecture—or a video podcast, if available—instead of going to class? After all, you hear and perhaps see the lecture just as if you were there, and you can sleep late and “go” to this class whenever it’s convenient for you. What could be wrong with that?
because you miss out on asking questions to other class mates and the instructor
-
Carla wants to get through college, and she knows she needs the degree to get a decent job, but she’s just not that into it. She’s never thought of herself as a good student, and that hasn’t changed much in college. She has trouble paying attention in those big lecture classes, which mostly seem pretty boring. She’s pretty sure she can pass all her courses, however, as long as she takes the time to study before tests. It doesn’t bother her to skip classes when she’s studying for a test in a different class or finishing a reading assignment she didn’t get around to earlier. She does make it through her freshman year with a passing grade in every class, even those she didn’t go to very often. Then she fails the midterm exam in her first sophomore class. Depressed, she skips the next couple classes, then feels guilty and goes to the next. It’s even harder to stay awake because now she has no idea what they’re talking about. It’s too late to drop the course, and even a hard night of studying before the final isn’t enough to pass the course. In two other classes, she just barely passes. She has no idea what classes to take next term and is starting to think that maybe she’ll drop out for now.
Carla is setting her self up for failure because she dose not go to class and relies on other for notes and cant ask questions.
-
Carla wants to get through college, and she knows she needs the degree to get a decent job, but she’s just not that into it. She’s never thought of herself as a good student, and that hasn’t changed much in college. She has trouble paying attention in those big lecture classes, which mostly seem pretty boring. She’s pretty sure she can pass all her courses, however, as long as she takes the time to study before tests. It doesn’t bother her to skip classes when she’s studying for a test in a different class or finishing a reading assignment she didn’t get around to earlier. She does make it through her freshman year with a passing grade in every class, even those she didn’t go to very often. Then she fails the midterm exam in her first sophomore class. Depressed, she skips the next couple classes, then feels guilty and goes to the next. It’s even harder to stay awake because now she has no idea what they’re talking about. It’s too late to drop the course, and even a hard night of studying before the final isn’t enough to pass the course. In two other classes, she just barely passes. She has no idea what classes to take next term and is starting to think that maybe she’ll drop out for now.
I think Carla is trying to do the "quicker way". But in reality it's not the fastest way it is in my opinion is the lazier way of going about things in a productive and efficient way.
-
-
social-media-ethics-automation.github.io social-media-ethics-automation.github.io
-
How do you think about the relationship between social media and “real life”?
In contrast to how people are saying "social media isn't real life," I think that people tend to be a more exaggerated version of themselves when engaging online. I think this might be because they don't feel an immediate consequence for poor or extreme behavior alongside the fact that it's very easy to just say whatever you want and click post. It's easy to post impulsive thoughts and not think about how others would react, especially when you're not physically seeing any effects in your real life. So in a way, I think that social media is a part of our real life, but in a much more exaggerated way now that social pressure is generally lightened.
-
-
slatestarcodex.com slatestarcodex.com
-
Consider a very formulaic conservative radio show. Every week, the host talks about some scandal that liberals have been involved in. Then she explains why it means the country is going to hell. I don’t think the listeners really care that a school in Vermont has banned Christmas decorations or whatever. The point is to convey this vague undercurrent of “Hey, there are other people out there who think like you, we all agree with you, you’re a good person, you can just sit here and listen and feel reassured that you’re right.” Anything vaguely conservative in content will be equally effective, regardless of whether the listener cares about the particular issue.
I've probably read this post half a dozen times over the last 10 years.
This is the first time that I've noticed (and—now—am able to appreciate) that this paragraph is deliberately crafted to, in part, have the same effect as what it's actually describing.
-
-
-
Author response:
The following is the authors’ response to the original reviews.
Public Reviews:
Reviewer #1 (Public Review):
Summary:
The manuscript submitted by Langenbacher et al., entitled " Rtf1-dependent transcriptional pausing regulates cardiogenesis", describes very interesting and highly impactful observations about the function of Rtf-1 in cardiac development. Over the last few years, the Chen lab has published novel insights into the genes involved in cardiac morphogenesis. Here, they used the mouse model, the zebrafish model, cellular assays, single cell transcription, chemical inhibition, and pathway analysis to provide a comprehensive view of Rtf1 in RNAPII (Pol2) transcription pausing during cardiac development. They also conducted knockdown-rescue experiments to dissect the functions of Rtf1 domains.
Strengths:
The most interesting discovery is the connection between Rtf1 and CDK9 in regulating Pol2 pausing as an essential step in normal heart development. The design and execution of these experiments also demonstrate a thorough approach to revealing a previously underappreciated role of Pol2 transcription pausing in cardiac development. This study also highlights the potential amelioration of related cardiac deficiencies using small molecule inhibitors against cyclin dependent kinases, many of which are already clinically approved, while many other specific inhibitors are at various preclinical stages of development for the treatment of other human diseases. Thus, this work is impactful and highly significant.
We thank the reviewer for appreciating our work.
Reviewer #2 (Public Review):
Summary:
Langenbacher at el. examine the requirement of Rtf1, a component of the PAF1C, which regulates transcriptional pausing in cardiac development. The authors first confirm their previous morphant study with newly generated rtf1 mutant alleles, which recapitulate the defects in cardiac progenitor and diUerentiation gene expression observed previously in morphants. They then examine the conservation of Rtf1 in mouse embryos and embryonic stem cell-derived cardiomyocytes. Conditional loss of Rtf1 in mesodermal lineages and depletion in murine ESCs demonstrates a failure to turn on cardiac progenitor and diUerentiation marker genes, supporting conservation of Rtf1 in promoting cardiac development. The authors subsequently employ bulk RNA-seq on flow-sorted hand2:GFP+ cells and multiomic single-cell RNA-seq on whole Rtf1-depleted embryos at the 10-12 stage. These experiments corroborate that genes associated with cardiac and muscle development are lost. Furthermore, the diUerentiation trajectories suggest that the expression of genes associated with cardiac maturation is not initiated. Structure-function analysis supports that the Plus3 domain is necessary for its function in promoting cardiac progenitor formation. ChIP-seq for RNA Pol II on 1012 somite stage embryos suggests that Rtf1 is required for proper promoter pausing. This defect can partially be rescued through use of a pharmacological inhibitor for Cdk9, which inhibits elongation, can partially restore elongation in rtf1 mutants.
Strengths:
Many aspects of the data are strong, which support the basic conclusions of the authors that Rtf1 is required for transcriptional pausing and has a conserved requirement in vertebrate cardiac development. Areas of strength include the genetic data supporting the conserved requirement for Rtf1 in promoting cardiac development, the complementary bulk and single-cell RNA-sequencing approaches providing some insight into the gene expression changes of the cardiac progenitors, the structure-function analysis supporting the requirement of the Plus3 domain, and the pharmacological epistasis combined with the RNA Pol II ChIP-seq, supporting the mechanism implicating Cdk9 in the Rtf1 dependent mechanism of RNA Pol II pausing.
We thank the reviewer for the summary and for recognizing many strengths of our work.
Weaknesses:
While most of the basic conclusions are supported by the data, there are a number of analyses that are confusing as to why they chose to perform the experiments the way they did and some places where the interpretations presently do not support the interpretations. One of the conclusions is that the phenotype aUects the maturation of the cardiomyocytes and they are arresting in an immature state. However, this seems to be mostly derived from picking a few candidates from the single cell data in Fig. 6. If that were the case, wouldn't the expectation be to observe relatively normal expression of earlier marker genes required for specification, such as Nkx2.5 and Gata5/6? The in situ expression analysis from fish and mice (Fig. 2 and Fig. 3) and bulk RNA-seq (Fig. 5) seems to suggest that there are pretty early specification and diUerentiation defects. While some genes associated with cardiac development are not changed, many of these are not specific to cardiomyocyte progenitors and expressed broadly throughout the ALPM. Similarly, it is not clear why a consistent set of cardiac progenitor genes (for instance mef2ca, nkx2.5, and tbx20) was analyzed for all the experiments, in particular with the single cell analysis.
A major conclusion of our study is that Rtf1 deficiency impairs myocardial lineage differentiation from mesoderm, as suggested by the reviewer. Thus, the main goal of this study is to understand how Rtf1 drives cardiac differentiation from the LPM, rather than the maturation of cardiomyocytes. Multiple lines of evidence support this conclusion:
(a) In situ hybridization showed that Rtf1 mutant embryos do not have nkx2.5+ cardiac progenitor cells and subsequently fail to produce cardiomyocytes (Figs. 2, 3).
(b) RT-PCR analysis showed that knockdown of Rtf1 in mouse embryonic stem cells causes a dramatic reduction of cardiac gene expression and production of significantly fewer beating patches (Fig.4).
(c) Bulk RNA sequencing revealed significant downregulation of cardiac lineage genes, including nkx2.5 (Fig. 5).
(d) Single cell RNA sequencing clearly showed that lateral plate mesoderm (LPM) cells are significantly more abundant in Rtf1 morphant,s whereas cardiac progenitors are less abundant (Fig. 6 and Fig.6 Supplement 1-5).
When feasible, we used cardiac lineage restricted markers in our assays. Nkx2.5 and tbx5a are not highlighted in the single cell analysis because their expression in our sc-seq dataset was too low to examine in the clustering/trajectory analysis. In this revised manuscript, we provide violin plots showing the low expression levels of these genes in single cells from Rtf1 deficient embryos (Figure 6 Supplement 5).
The point of the multiomic analysis is confusing. RNA- and ATAC-seq were apparently done at the same time. Yet, the focus of the analysis that is presented is on a small part of the RNA-seq data. This data set could have been more thoroughly analyzed, particularly in light of how chromatin changes may be associated with the transcriptional pausing. This seems to be a lost opportunity. Additionally, how the single cell data is covered in Supplemental Fig. 2 and 3 is confusing. There is no indication of what the diUerent clusters are in the Figure or the legend.
In this study, we performed single cell multiome analysis and used both scRNAseq and scATACseq datasets to generate reliable clustering. The scRNAseq analysis reveals how Rtf1 deficiency impacts cardiac differentiation from mesoderm, which inspired us to investigate the underlying mechanism and led to the discovery of defects in Rtf1-dependent transcriptional pause release.
We agree with the reviewer that deep examination of Rtf1-dependent chromatin changes would provide additional insights into how Rtf1 influences early development and careful examination of the scATACseq dataset is certainly a good future direction.
In this revised manuscript, we have revised Fig.6 Supplement 1 to include the predicted cell types and provide an additional excel file showing the annotation of all 39 clusters (Supplementary Table 2).
While the effect of Rtf1 loss on cardiomyocyte markers is certainly dramatic, it is not clear how well the mutant fish have been analyzed and how specific the eUect is to this population. It is interpreted that the eUects on cardiomyocytes are not due to "transfating" of other cell fates, yet supplemental Fig. 4 shows numerous eUects on potentially adjacent cell populations. Minimally, additional data needs to be provided showing the live fish at these stages and marker analysis to support these statements. In some images, it is not clear the embryos are the same stage (one can see pigmentation in the eyes of controls that is not in the mutants/morphants), causing some concern about developmental delay in the mutants.
Single cell RNA sequencing showed an increased abundance of LPM cells and a reduced abundance of cardiac progenitors in Rtf1 morphants (Fig. 6 and Fig.6 Supplement 1-5). The reclustering of anterior lateral plate mesoderm (ALPM) cells and their derivatives further showed that cells representing undifferentiated ALPM were increased whereas cells representing all three ALPM derivatives were reduced. These findings indicate a defect in ALPM differentiation.
The reviewer questioned whether we examined stage-matched embryos. In our assay, Rtf1 mutant embryos were collected from crosses of Rtf1 heterozygotes. Each clutch from these crosses consists of ¼ embryos showing rtf1 mutant phenotypes and ¾ embryos showing wild type phenotypes which were used as control. Mutants and their wild type siblings were fixed or analyzed at the same time.
The reviewer questioned the specificity of the Rtf1 deficient cardiac phenotype and pointed out that Rtf1 mutant embryos do not have pigment cells around the eye. Rtf1 is a ubiquitously expressed transcriptional regulator. Previous studies in zebrafish have shown that Rtf1 deficiency significantly impacts embryonic development. Rtf1 deficiency causes severe defects in cardiac lineage and neural crest cell development; consequently, Rtf1 deficient embryos do not have cardiomyocytes and pigmentation (Langenbacher et al., 2011, Akanuma et al., 2007, and Jurynec et al., 2019). We now provide an image showing a 2-day-old Rtf1 mutant embryo and their wild type sibling to illustrate the cardiac, neural crest, and somitogenesis defects caused by loss of Rtf1 activity (Fig. 2 Supplement 1).
With respect to the transcriptional pausing defects in the Rtf1 deficient embryos, it is not clear from the data how this eUect relates to the expression of the cardiac markers. This could have been directly analyzed with some additional sequencing, such as PRO-seq, which would provide a direct analysis of transcriptional elongation.
We showed that Rtf1 deficiency results in a nearly genome-wide decrease in promoterproximal pausing and downregulation of cardiac makers. Attenuating transcriptional pause release could restore cardiomyocyte formation in Rtf1 deficient embryos. In this revised manuscript, we provide additional RNAseq data showing that the expression levels of critical cardiac development genes such as nkx2.5, tbx5a, tbx20, mef2ca, mef2cb, ttn.2, and ryr2b are significantly rescued. We agree with the reviewer that further analyses using the PRO-seq approach could provide additional insights, but it is beyond the scope of this manuscript.
Some additional minor issues include the rationale that sequence conservation suggests an important requirement of a gene (line 137), which there are many examples this isn't the case, referencing figures panels out of order in Figs. 4, 7, and 8) as described in the text, and using the morphants for some experiments, such as the rescue, that could have been done in a blinded manner with the mutants.
We have clarified the rationale in this revised manuscript and made the eRort to reference figures in order.
The reviewer commented that rescue experiments “could have been done in a blinded manner with the mutants”. This was indeed how the flavopiridol rescue and cdk9 knockdown experiments were carried out. Embryos from crosses of Rtf1 heterozygotes were collected, fixed after treatment and subjected to in situ hybridization. Embryos were then scored for cardiac phenotype and genotyped (Fig.8 d-g). Morpholino knockdown was used in genomic experiments because our characterization of rtf1 morphants showed that they faithfully recapitulate the rtf1 mutant phenotype during the timeframe of interest (Fig. 2).
Recommendations for the authors:
Reviewer #1 (Recommendations For The Authors):
This reviewer has a few suggestions below, aimed at improving the clarity and impact of the current study. Once these items are addressed, the manuscript should be of interest to the Elife reader.
Item 1. Strengthening the interaction between Rfh1 and CDK9 on Pol2 pausing.
The authors have convincingly shown that the chemical inhibition of CDK9 by flavopiridol can partially rescue the expression of cardiac genes in the zebrafish model. Although flavopiridol is FDA approved and has been a classical inhibitor for the dissection of CDK9 function, it also inhibits related CDKs (such as Flavopiridol (Alvocidib) competes with ATP to inhibit CDKs including CDK1, CDK2, CDK4, CDK6, and CDK9 with IC50 values in the 20-100 nM range) Therefore, this study could be more impactful if the authors can provide evidence on which of these CDKs may be most relevant during Rtf1-dependent cardiogenesis. To determine whether the observed cardiac defect indicates a preferential role for CDK9, or that other CDKs may also be able to provide partial rescue may be clarified using additional, more selective small molecules (e.g., BAY1251152, LDC000067 are commercially available).
The reviewer raised a reasonable concern about the specificity of flavopiridol. We thank the reviewer for the insightful suggestion and share the concern about specificity. To address this question, we have used an orthogonal testing through morpholino inhibition where we directly targeted CDK9 and observed the same level of rescue, supporting a critical role of transcription pausing in cardiogenesis.
Item 2. Differences between CRISPR lines and morphants
Much of the work presented used Rtf1 morphants while the authors have already generated 2 CRISPR lines. What is the diUerence between morphants and mutants? The authors should comment on the similarities and/or differences between using morphants or mutants in their study and whether the same Rtf1- CDK9 connection also occurs in the CRISPR lines.
The morphology of our mutants (rtf1<sup>LA2678</sup> and rtf1<sup>LA2679</sup>) resembles the morphants and the previously reported ENU-induced rtf1<sup>KT641</sup> allele. Extensive in situ hybridization analysis showed that the morphants faithfully recapitulate the mutant phenotypes (Fig.2). We have performed rescue experiments (flavopiridol and CDK9 morpholino) using Rtf1 mutant embryos and found that inhibiting Cdk9 restores cardiomyocyte formation (Fig.8).
Item 3. Discuss the therapeutic relevance of study
The authors have already generated a mouse model of Rtf1 Mesp1-Cre knockout where cardiac muscle development is severely derailed (Fig 3B). Thus, a demonstration of a conserved role for CDK9 inhibitor in rescuing cardiogenesis using mouse cells or the mouse model will provide important information on a conserved pathway function relevant to mammalian heart development. In the Discussion, how this underlying mechanistic role may be useful in the treatment of congenital heart disease should be provided.
Thank you for the insight. We have incorporated your comments in the discussion.
Item 4. Insights into the role of CDK9-Rtf1 in response to stress versus in cardiogenesis.
In the Discussion, the authors commented on the role of additional stress-related stimuli such as heat shock and inflammation that have been linked to CDK9 activity. However, the current ms provides the first, endogenous role of Pol2 pausing in a critical developmental step during normal cardiogenesis. The authors should emphasize the novelty and significance of their work by providing a paragraph on the state of knowledge on the molecular mechanisms governing cardiogenesis, then placing their discovery within this framework. This minor addition will also clarify the significance of this work to the broad readership of eLife.
Thank you for the suggestion. We have incorporated your comments and elaborate on the novelty and significance of our work in the discussion.
Reviewer #2 (Recommendations For The Authors):
(1) It is diUicult to assess what the overt defects are in the embryos at any stages. Images of live images were not included in the supplement. Do these have a small, malformed heart tube later or are the embryos just deteriorating due to broad defects?
The Rtf1 deficient embryos do not produce nkx2.5+ cardiac progenitors. Consequently, we never observed a heart tube or detected cells expressing cardiomyocyte marker genes such as myl7. This finding is consistent with previous reports using rtf1 morphants and rtf<sup>1KT64</sup>, an ENU-induced point mutation allele (Langenbacher et al., 2011 and Akanuma, 2007). In this revised manuscript, we provide a live image of 2-day-old wild type and rtf1<sup>LA2679/LA2679</sup> embryos (Fig. 2 Supplement 1). After two days, rtf1 mutant embryos undergo broad cell death.
(2) Fig. 2, although the in situs are convincing, there is not a quantitative assessment of expression changes for these genes. This could have been done for the bulk or single cell RNA-seq experiments, but was not and these genes weren't not included in the heat maps. A quantitative assessment of these genes would benefit the study.
The top 40 most significantly differentially expressed genes are displayed in the heatmap presented in Fig.5d. The complete differential gene expression analysis results for our hand2 FACS-based comparison of rtf1 morphants and controls is presented in Supplementary Data File 1. In this revised manuscript, we provide a new supplemental figure with violin plots showing the expression levels of genes of interest in our single cell sequencing dataset (Fig.6 Supplement 5).
(3) It doesn't not appear that any statistical tests were used for the comparisons in Fig. 2.
We now provide the statistical data in the legend and Fig.2 b, d, f, h and i.
(4) It's not clear the magnifications and orientations of the embryos in Fig. 3b are the same.
Embryos shown in Fig.3b are at the same magnification. However, because Rtf1 mutant embryos display severe morphological defects, the orientation of mutant embryos was adjusted to examine the cardiac tissue.
(5) The n's for analysis of MLC2v in WT Rtf1 CKO embryos in Fig. 3b are only 1. At least a few more embryos should be analyzed to confirm that the phenotype is consistent.
We have revised the figure and present the number of embryos analyzed and statistics in Fig.3c.
(6) A number of figure panels are referred to out of order in the text. Fig. 4E-G are before Fig. 4C, D, Fig. 7C before 7B, Fig. 8D-I before 8A ,B. In general, it is easier for the reader if the figures panels are presented in the order they are referred to in the text.
Revised as suggested.
(7) While additional genes can be included, it is not clear why the same sets of genes are not examined in the bulk or single-cell RNA-seq as with the in situs or expression was analyzed in embryos. I suggest including the genes like nkx2.5, tbx20, myl7, in all the sequencing analysis.
We used the same set of genes in all analyses when possible. However, the low expression of genes such as nkx2.5 and myl7 in our sc-seq dataset preclude them from the clustering/trajectory analysis. In this revised manuscript, we present violin plots showing their expression in wild type and rtf1 morphants (Fig. 6 Supplement 5).
(8) If a multiomic approach was used, why wasn't its analysis incorporated more into the manuscript? In general, a clearer presentation and deeper analysis of the single cell data would benefit the study. The integration of the RNA and ATAC would benefit the analysis.
As addressed in our response to the reviewer’s public review, both datasets were used in clustering. Examining changes in chromatin accessibility is certainly interesting, but beyond the scope of this study.
(9) Many of the markers analyzed are not cardiac specific or it is not clear they are expressed in cardiac progenitors at the stage of the analysis. Hand2 has broader expression. Additional confirmation of some of the genes through in situ would help the interpretations.
Markers used for the in situ hybridization analysis (myl7, mef2ca, nkx2.5, tbx5a, and tbx20) are known for their critical role in heart development. For sc-seq trajectory analyses, most displayed genes (sema3e, bmp6, ttn.2, mef2cb, tnnt2a, ryr2b, and myh7bb) were identified based on their differential expression along the LPM-cardiac progenitor pseudotime trajectory. Rather than selecting genes based on their cardiac specificity, our goal was to examine the progressive gene expression changes associated with cardiac progenitor formation and compare gene expression of wild type and rtf1 deficient embryos.
(10) Additional labels of the cell clusters are needed for Supplemental Figs. 2 and 3.
The cluster IDs were presented on Supplementary Figures 2 and 3. In this revised version, we added predicted cell types to the UMAP (revised Fig.6 Supplement 1) and provided an excel file with this information (revised Supplementary Table 2).
(11) On lines 101-102, the interpretation from the previous data is that diUerentiation of the LPM requires Rtf1. However, later from the single cell data the interpretation based on the markers is that Rtf1 loss aUects maturation. However, it is not clear this interpretation is correct or what changed from the single cell data. If that were the case, one would expect to see maintenance of more early marks and subsequent loss of maturation markers, which does not appear to the be the case from the presented data.
Our data suggests that cardiac progenitor formation is not accomplished by simultaneously switching on all cardiac marker genes. Our pseudotime trajectory analysis highlights tnnt2a, ryr2b, and myh7bb as genes that increase in expression in a lagged manner compared to mef2cb (Fig. 6). Thus, the abnormal activation of mef2cb without subsequent upregulation of tnnt2a, ryr2b, and myh7bb in rtf1 morphants suggests a requirement for rtf1 in the progressive gene expression changes required for proper cardiac progenitor differentiation. Our single cell experiment focuses on the process of cardiac progenitor differentiation and does not provide insights into cardiomyocyte maturation. We have edited the text to clarify these interpretations.
(12) The interpretation that there is not "transfating" is not supported by the shown data. Analysis of markers in other tissues, again with in situ, to show spatially would benefit the study.
As stated in our response to the reviewer’s public review, we observed a dramatic increase of ALPM cells, but a decrease of ALPM derivatives including the cardiac lineage. We did not observe the expansion of one ALPM-derived subpopulation at the expense of the others. These observations suggest a defect in ALPM differentiation and argue against the notion that the region of the ALPM that would normally give rise to cardiac progenitors is instead differentiating into another cell type.
(13) The rationale that sequence conservation means a gene is important (lines 137-139) is not really true. There are examples a lot of highly conserved genes whose mutants don't have defects.
We have revised the text to avoid confusion.
(14) The data showing that the 8 bp mutations do not aUect the RNA transcript is not shown or at least indicated in Fig. 7. It would seem that this experiment could have been done in the mutant embryos, in which case the experiment would have been semi-blinded as the genotyping would occur after imaging.
The modified Rtf1 wt RNA (Rtf1 wt* in revised Fig. 7) robustly rescued nkx2.5 expression in rtf1 deficient embryos, demonstrating that the 8 bp modifications do not negatively impact the activity of the injected RNA. As stated previously, morpholino knockdown was used in some experiments because our characterization of rtf1 morphants showed that they faithfully recapitulate the rtf1 mutant phenotype during the timeframe of interest.
(15) Using a technique like PRO-seq at the same stage as the ChIP-seq would complement the ChIP-seq and allow a more detailed analysis of the transcriptional pausing on specific genes observed in WT and mutant embryos.
As stated in our response to the reviewer’s public review, we appreciate the suggestion but PRO-seq is beyond the scope of this study.
-
-
santafe.edu santafe.edu
-
Describing Anderson’s approach to physics, Yu recalled him saying, “Theoretical physics is not just doing calculations. It's setting up the problem so that any fool could do the calculation.”
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the previous reviews.
Public Reviews:
Reviewer #1 (Public review):
The authors assess the impact of E-cigarette smoke exposure on mouse lungs using single cell RNA sequencing. Air was used as control and several flavors (fruit, menthol, tobacco) were tested. Differentially expressed genes (DEGs) were identified for each group and compared against the air control. Changes in gene expression in either myeloid or lymphoid cells were identified for each flavor and the results varied by sex. The scRNAseq dataset will be of interest to the lung immunity and e-cig research communities and some of the observed effects could be important. Unfortunately, the revision did not address the reviewers' main concerns about low replicate numbers and lack of validations. The study remains preliminary, and no solid conclusions could be drawn about the effects of E-cig exposure as a whole or any flavor-specific phenotypes.
Strengths:
The study is the first to use scRNAseq to systematically analyze the impact of e-cigarettes on the lung. The dataset will be of broad interest.
Weaknesses:
scRNAseq studies may have low replicate numbers due to the high cost of studies but at least 2 or 3 biological replicates for each experimental group is required to ensure rigor of the interpretation. This study had only N=1 per sex per group and some sex-dependent effects were observed. This could have been remedied by validating key observations from the study using traditional methods such as flow cytometry and qPCR, but the limited number of validation experiments did not support the conclusions of the scRNA seq analysis. An important control group (PG:VG) had extremely low cell numbers and was basically not useful. Statistical analysis is lacking in almost all figures. Overall, this is a preliminary study with some potentially interesting observations, but no solid conclusions can be made from the data presented.
The only new validation experiment is the immunofluorescent staining of neutrophils in Figure 4. The images are very low resolution and low quality and it is not clear which cells are neutrophils. S100A8 (calprotectin) is highly abundant in neutrophils but not strictly neutrophil-specific. It's hard to distinguish positive cells from autofluorescence in both Ly6g and S100a8 channels. No statistical analysis in the quantification.
We thank the reviewer for identifying the strengths of this study and pointing out the gaps in knowledge. Overall, our purpose to present this data is to provide the scRNA seq results as a resource to a wider community. We have used techniques like flow cytometry, multianalyte cytokine array and immunofluorescence to validate some of the results. We agree with the reviewer that we were unable to rightly point out the significance of our findings with the immunofluorescent stain in the previous edit. We have revised the manuscript and included the quantification for both Ly6G+ and S100A8+ cells in e-cig aerosol exposed and control lung tissues. Briefly, we identified a marked decrease in the staining for S100A8 (marker for neutrophil activation) in tobacco-flavored e-cig exposed mouse lungs as compared to controls. Upon considering the corroborating evidence from scRNA seq and flow cytometry with regards to increased neutrophil percentages in experimental group and lowered staining for active neutrophils using immunofluorescence, we speculate that exposure to e-cig (tobacco) aerosols may alter the neutrophil dynamics within the lungs. Also, co-immunofluorescence identified a more prominent co-localization of the two markers in control samples as compared to the treatment group which points towards some changes in the innate immune milieu within the lungs upon exposures. Future work is required to validate these speculations.
We have now discussed all the above-mentioned points in the Discussion section of the revised manuscript and toned down our conclusions regarding sex-dependent changes from scRNA seq data.
It is unclear what the meaning of Fig. 3A and B is, since these numbers only reflect the number of cells captured in the scRNAseq experiment and are not biologically meaningful. Flow cytometry quantification is presented as cell counts, but the percentage of cells from the CD45+ gate should be shown. No statistical analysis is shown, and flow cytometry results do not support the conclusions of scRNAseq data.
We thank the reviewer for this question. However, we would like to highlight that scRNA seq and flow cytometry may show similar trends but cannot be identical as one relies on cell surface markers (protein) for identification of cell types, while other is dependent on the transcriptomic signatures to identify the cell types. In our data, for the myeloid cells (alveolar macrophages and neutrophils), the scRNA and flow cytometry data match in trend. However, the trends do not match with respect to the lymphoid cells being studied (CD4 and CD8 T cells). The possible explanation for such a finding could be possible high gene dropout rates in scRNA seq, different analytical resolution for the two techniques and pooling of samples in our single cell workflow. We realize these shortcomings in our analyses and mention it clearly in the discussion as limitation of our work. It is important to note also that cell frequencies identified in scRNA seq just provide wide and indistinct indications which need to be further validated, which we tried to accomplish in our work to some degree. Our flow-based results clearly highlight the sex-specific variations in the immune cell percentages (something we could not have anticipated earlier). In future studies, we will include more replicates to tease out sex-based variations upon acute and chronic exposure to e-cig aerosols.
We have now replotted the graphs in Fig 3A and B and plotted the flow quantification as the percentage of total CD45+ cells. The gating strategy for the flow plots is also included as Figure S6 in the revised manuscript.
Reviewer #2 (Public review):
This study provides some interesting observations on how different flavour e-cigarettes can affect lung immunology; however, there are numerous flaws, including a low replicate number and a lack of effective validation methods, meaning findings may not be repeated. This is a revised article but several weaknesses remain related to the analysis and interpretation of the data.
Strengths:
The strength of the study is the successful scRNA-seq experiment which gives some preliminary data that can be used to create new hypotheses in this area.
Weaknesses:
Although some text weaknesses have been addressed since resubmission, other specific weaknesses remain: The major weakness is the n-number and analysis methods. Two biological n per group is not acceptable to base any solid conclusions. Any validatory data was too little (only cell % data) and not always supporting the findings (e.g. figure 3D does not match 3B/4A). Other examples include:
There aren't enough cells to justify analysis - only 300-1500 myeloid cells per group with not many of these being neutrophils or the apparent 'Ly6G- neutrophils'.
We thank the reviewer for the comment, but we disagree with the reviewer in terms of the justification of analyses. All the flavored e-cig aerosol groups were compared with air controls to deduce the outcomes in the current study. We already acknowledge low sample quality for PGVG group and have only included the comparisons with PGVG upon reviewer’s request which is open to interpretation by the reader.
By that measure, each treatment group (except PGVG group) has over 1000 cells with 24777 genes being analyzed for each cell type, which by the standards of single cell is sufficient. We understand that this strategy should not be used for detection of rare cell populations, which was neither the purpose of this manuscript nor was attempted. We conduct comparisons of broader cell types and mention more samples need to be added in the Discussion section of the revised manuscript.
As for the Ly6G neutrophil category, we don’t only base our results on scRNA analyses but also perform co-immunofluorescence and multi-analyte analyses and use evidence from previous literature to back our outcome. To avoid over-stating our results we have revamped the whole manuscript and ensured to tone down our results with relation to the presence of Ly6G- neutrophils. We do understand that more work is required in the future, but our work clearly shows the shift in neutrophil dynamics upon exposure which should be reported, in our opinion.
The dynamic range of RNA measurement using scRNAseq is known to be limited - how do we know whether genes are not expressed or just didn't hit detection? This links into the Ly6G negative neutrophil comments, but in general the lack of gene expression in this kind of data should be viewed with caution, especially with a low n number and few cells. The data in the entire paper is not strong enough to base any solid conclusion - it is not just the RNA-sequencing data.
We acknowledge this to be a valid point and have revamped the manuscript and toned down our conclusions. However, such limitations exist with any scRNA seq dataset and so must be interpreted accordingly by the readers. We do understand that due to the low cell counts and the limitations with scRNA seq we should not perform DESeq2 analyses for Ly6G+ versus Ly6G- neutrophil categories, which was never attempted at the first place. However, our results with co-immunofluorescence, multianalyte assay and scRNA expression analyses in myeloid cluster do point towards a shift in neutrophil activation which needs to be further investigated. Furthermore, Ly6G deficiency has been linked to immature neutrophils in many previous studies and is not an unlikely outcome that needs to be treated with immense skepticism.
We wish to make this dataset available as a resource to influence future research. We are aware of its limitations and have been transparent with regards to our experimental design, capture strategy, the quality of obtained results, and possible caveats to make it is open for discussion by the readers.
There is no data supporting the presence of Ly6G negative neutrophils. In the flow cytometry only Ly6G+ cells are shown with no evidence of Ly6G negative neutrophils (assuming equal CD11b expression). There is no new data to support this claim since resubmission and the New figures 4C and D actually show there are no Ly6G negative cells - the cells that the authors deem Ly6G negative are actually positive - but the red overlay of S100A8 is so strong it blocks out the green signal - looking to the Ly6G single stains (green only) you can see that the reported S100A8+Ly6G- cells all have Ly6G (with different staining intensities).
We thank the reviewer for this query and do understand the skepticism. We have now quantified the data to provide more clarity for interpretation. As we were using paraffin embedded tissues, some autofluorescence is expected which could explain some of reviewer’s concerns. However we expect that the inclusion of better quality images and quantification must address some of the concerns raised by the reviewer.
Eosinophils are heavily involved in lung macrophage biology, but are missing from the analysis - it is highly likely the RNA-sequence picked out eosinophils as Ly6G- neutrophils rather than 'digestion issues' the authors claim
We thank the reviewer for raising a valid concern. However, the Ly6G- cluster cannot be eosinophils in our case. Literature suggests SiglecF as an important biomarker of eosinophils which was absent in the Ly6G- cluster our in scRNA seq analyses as shown in File S18 and Figure 6B of the revised manuscript. We have now provided a detailed explanation (Lines 476-488; 503-506) of the observed results pertaining to eosinophil population in the revised manuscript to further address some of the concerns raised by this reviewer.
After author comments, it appears the schematic in Figure 1A is misleading and there are not n=2/group/sex but actually only n=1/group/sex (as shown in Figure 6A). Meaning the n number is even lower than the previous assumption.
We concur with reviewers’ valid concern and so are willing to provide this data as a resource for a wider audience to assist future work. Pooling of samples have been practiced by many groups previously to save resources and expense. We did it for the very same reason. It may not be the preferred approach, but it still has its merit considering the vast amount of cell-specific data generated using this strategy. To avoid overstating our results we have ensured to maintain transparency in our reporting and acknowledge all the limitations of this study.
We do not believe that the strength of scRNA seq lies in drawing conclusive results, but to tease our possible targets and direction that need to be validated with more work. In that respect, our study does identify the target cell types and biological processes which could be of importance for future studies.
Reviewer #3 (Public review):
This work aims to establish cell-type specific changes in gene expression upon exposure to different flavors of commercial e-cigarette aerosols compared to control or vehicle. Kaur et al. conclude that immune cells are most affected, with the greatest dysregulation found in myeloid cells exposed to tobacco-flavored e-cigs and lymphoid cells exposed to fruit-flavored e-cigs. The up- and down-regulated genes are heavily associated with innate immune response. The authors suggest that a Ly6G-deficient subset of neutrophils is found to be increased in abundance for the treatment groups, while gene expression remains consistent, which could indicate impaired function. Increased expression of CD4+ and CD8+ T cells along with their associated markers for proliferation and cytotoxicity is thought to be a result of activation following this decline in neutrophil-mediated immune response.
Strengths:
Single cell sequencing data can be very valuable in identifying potential health risks and clinical pathologies of lung conditions associated with e-cigarettes considering they are still relatively new.
Not many studies have been performed on cell-type specific differential gene expression following exposure to e-cig aerosols.
The assays performed address several factors of e-cig exposure such as metal concentration in the liquid and condensate, coil composition, cotinine/nicotine levels in serum and the product itself, cell types affected, which genes are up- or down-regulated and what pathways they control.
Considerations were made to ensure clinical relevance such as selecting mice whose ages corresponded with human adolescents so that data collected was relevant.
Weaknesses:
The exposure period of 1 hour a day for 5 days is not representative of chronic use and this time point may be too short to see a full response in all cell types. The experimental design is not well-supported based on the literature available for similar mouse models. Clinical relevance of this short exposure remains unclear.
We thank the reviewer for this query. However, we would like to emphasize that chronic exposure was never the intention of this study. We wished to design a study for acute nose-only exposure owing to which the study duration was left shorter. Shorter durations limit the stress and discomfort to the animal. The in vivo study using nose-only exposure regimen is still developing with multiple exposure regimen being used by different groups. To our knowledge there is no gold standard of e-cig aerosol exposure which is widely accepted other than the CORESTA recommendations, which we followed. Also, we show in our study how the daily exposure to leached metals vary in a flavor-dependent manner thus validating that exposure regime does need more attention in terms of equal dosing, particle distribution and composition- something we have started doing in our future studies. We have included all the explanations in the revised manuscript (Lines 82-85, 425-435, 648-654).
Several claims lack supporting evidence or use data that is not statistically significant. In particular, there were no statistical analyses to compare results across sex, so conclusions stating there is a sex bias for things like Ly6G+ neutrophil percentage by condition are observational.
We agree with reviewer’s comment and have taken this into consideration. We have now revamped the whole manuscript and toned down most of the sex-based conclusions stated in this work. Having said that, it is important to note that most of the work relying solely on scRNA seq, as is the case for this study, is observational in nature and needs to be assessed bearing this in mind.
Overall, the paper and its discussion are relatively surface-level and do not delve into the significance of the findings or how they fit into the bigger picture of the field. It is not clear whether this paper is intended to be used as a resource for other researchers or as an original research article.
We have now reworked on the Discussion and tried to incorporate more in-depth discussion and the results providing our insights regarding the observations, discrepancies and the possible explanations. We have also made it clear that this paper is intended to be used as a resource by other researchers (Lines 577-579)
The manuscript has some validation of findings but not very comprehensive.
We have now revamped the manuscript. We have Included quantification for immunofluorescence data with better representation of the GO analyses. We have worked on the Results and Discussion sections to make this a useful resource for the scientific community.
This paper provides a strong foundation for follow-up experiments that take a closer look at the effects of e-cig exposure on innate immunity. There is still room to elaborate on the differential gene expression within and between various cell types.
We thank the reviewer for pointing out the strength of this paper. The reason why we refrained from elaborating of the differential gene expressions within and between various cell types was due to low sample number and sequencing depth for this study. However the raw data will be provided with the final publication, which should be freely accessible to the public to re-analyze the data set as they deem fit.
Comments on revisions:
The reviewers have addressed major concerns with better validation of data and improved organization of the paper. However, we still have some concerns and suggestions pertaining to the statistical analyses and justifications for experimental design.
We appreciate the nuance of this experimental design, and the reviewers have adequately commented on why they chose nose-only exposure over whole body exposure. However, the justification for the duration of the exposure, and the clinical relevance of a short exposure, have not been addressed in the revised manuscript.
We thank the editor for this query. We have now addressed this query briefly in Lines 82-85, 425-435, 648-654 of the revised manuscript. We would like to add, however, that we intend to design a study for acute nose-only exposure for this project. Shorter durations limit the stress and discomfort to the animal, owing to which a duration of 1hour per day was chosen. The in vivo study using nose-only exposure regimen is still developing with multiple exposure regimen being used by different groups. Ours is one such study in that direction just intended to identify cell-specific changes upon exposure. Considering our results in Figure 1B showing variations in the level of metals leached in each flavor per day, the appropriate exposure regimen to design a controlled, reproducible experiment needs to be discussed. There could be room for improvement in our strategy, but this was the best regimen that we found to be appropriate per the literature and our prior knowledge in the field.
The presentation of cell counts should be represented by a percentage/proportion rather than a raw number of cells. Without normalization to the total number of cells, comparisons cannot be made across groups/conditions. This comment applies to several figures.
We thank the editor for this comment and have now made the requested change in the revised manuscript.
We appreciate that the authors have taken the reviewers' advice to validate their findings. However, we have concerns regarding the immunofluorescent staining shown in Figure 4. If the red channel is showing a pan-neutrophil marker (S100A8) and the green channel is showing only a subset of neutrophils (LY6G+), then the green channel should have far less signal than the red channel. This expected pattern is not what is shown in the figure, with the Ly6G marker apparently showing more expression than S100A8. Additionally, the FACS data states that only 4-5% of cells are neutrophils, but the red channel co-localizes with far more than 4-5% of the DAPI stain, meaning this population is overrepresented, potentially due to background fluorescence (noise). In addition, some of the shapes in the staining pattern do not look like true neutrophils, although it is difficult to tell because there remains a lot of background staining. The authors need to verify that their S100A8 and Ly6G antibodies work and are specific to the populations they intend to target. It is possible that only the brightest spots are truly S100A8+ or Ly6G+.
We thank the editor for this comment and acknowledge that we may have made broad generalizations in our interpretation of our data previously. We have now revisited the data and quantified the two fluorescence for better interpretation of our results. We have also reassessed our conclusions from this data and reworded the manuscript accordingly. Briefly we believe that Ly6G deficiency could be an indication of the presence of immature neutrophils in the lungs. This is a common process of neutrophil maturation. An active neutrophil population has Ly6G and should also express S100A8 indicating a normal neutrophilic response against stressors. However, our results, despite some autofluorescence which is common with lung tissues, shows a marked decline in the S100A8+ cells in the lung of tobacco-flavored e-cig aerosol exposed mice as compared to air controls. We also do not see prominent co-localization of the two markers in exposed group thus proving a shift in neutrophil dynamics which requires further investigation. We would also like to mention here that S100A8 is predominantly expressed in neutrophils, but is also expressed by monocytes and macrophages, so that could explain the over-representation of these cells in our immunofluorescence results. We have now included this in the Discussion section (Lines 489- 538) of the revised manuscript.
Paraffin sections do not always yield the best immunostaining results and the images themselves are low magnification and low resolution.
We agree with the editor that paraffin sections may not yield best results, we have worked on the final figure to improve the quality of the displayed results and zoomed-in some parts of the merged image to show the differences in the co-localization patterns for the two markers in our treated and control groups for easier interpretation.
Please change the scale bars to white so they are more visible in each channel.
The merged image in Figure 6C now has a white scale bar.
We appreciate that this is a preliminary test used as a resource for the community, but there is interesting biology regarding immune cells that warrants DEG analysis by the authors. This computational analysis can be easily added with no additional experiments required.
We thank the editor for this comment and agree that interesting biology regarding immune cells could be explored upon performing the DEG analyses on individual immune populations. However, due to the small sample size, low sequencing depth and pooling of same sex animals in each treatment group, we refrained from performing that analyses fearing over-representation of our results. We will be providing the link to the raw data with this publication which will be freely accessible to public on NIH GEO resource to allow further analyses on this dataset by the judgement of the investigator who utilizes it as a resource.
Recommendations for the authors:
Reviewer #1 (Recommendations for the authors):
(Minor) The pathway analyses in Fig. 6-8 have different fonts than what's used in all other figures.
We have now made the requested change in the revised manuscript.
-
-
danwang.co danwang.co
-
China’s automotive success is biting into Germany more than anywhere else. I keep a scrapbook filled with mournful remarks that German executives offer to newspapers. “Most of what German Mittelstand firms do these days, Chinese companies can do just as well,” said a consultant to the Financial Times. “In my sector they look at the price-point of the market leader and sell for roughly half of that,” the boss of a medical devicemaker told the Economist. It’s never hard to find parades of gloomy Germans. Now more than ever it looks like their core competences are threatened by Chinese firms.
I see this too. But it's a weird paragraph. Yes the automotive industry is behaving like dinosaurs in Germany, but the two examples (Mittelstand is not the automotive industry, and a medical device maker) don't connect to the rest.
-
it’s not obvious that the US will have a monopoly on this technology, just as it could not keep it over the bomb.
compares AI dev and attempts to keep it for oneself to the dev of atomic bombs and containment
Tags
Annotators
URL
-
-
-
22:48 "It's the gravitas of the situation that I see, that frustrates me that other people don't see it. I have been "preparing" intentionally since Hurricane Katrina, but I grew up on a farm in upstate New York. I know how to hunt. I know how to butcher animals. I know how to grow food. I know row crops and gardens. I know foraging in the woods. I know how to fish and where to get water from. And I understand how to move in a rural environment, not just the topographical terrain, but the human terrain as well. Been doing that my whole life. One could say, I've been prepared for this by the hand of the Most High my whole life. And I I see it. I see it coming. And it... while I would love to be wrong, it bothers me that others who do see it, or pay lip service to seeing it, don't take it as seriously as they should."
preach. there is too much demoralization everywhere.
Tags
Annotators
URL
-
-
cghuisunshine.github.io cghuisunshine.github.io
-
Caribou
English (thorough explanation)
1) What “Caribou” literally means
Caribou is the common name for a large, hoofed, deer-like animal in the species Rangifer tarandus. In many parts of the world, the same species is called reindeer.
So in simple terms: Caribou = a large northern deer, closely related to (and often the same as) reindeer.
2) Caribou vs. reindeer (why two names?)
- “Caribou” is used mainly in North America, especially Canada and Alaska, and often for wild populations.
- “Reindeer” is used more in Europe and Asia, and it often refers to domesticated (herded) populations—though biologically they’re the same species.
Think of it like a naming convention:
- Caribou (North American / wild context)
- Reindeer (Eurasian / domesticated or general context)
3) Key features (what makes a caribou distinctive)
- Antlers: Both males and females can grow antlers (this is unusual among deer).
- Hooves: Wide hooves act like snowshoes, helping them walk on snow and soft ground.
- Cold adaptation: Thick fur and an efficient metabolism help them survive Arctic/subarctic winters.
- Migration: Many populations travel in large seasonal migrations.
4) Grammar: singular/plural form
Caribou is commonly used as both singular and plural:
- “I saw a caribou.” (one)
- “I saw three caribou.” (more than one)
Sometimes you’ll see caribous, but caribou is more common.
5) How the word might be used if it appears alone
If the excerpt is just “Caribou” by itself (like your example), it most likely serves as:
- a topic heading (a section about the animal),
- a label (e.g., a picture caption),
- a category name (wildlife list), or
- a symbolic reference (e.g., representing the North, wilderness, migration, resilience).
6) Pronunciation
- CARE-ih-boo (common English pronunciation) IPA often shown as /ˈkærɪbuː/
中文(详细解释)
1)“Caribou”的基本意思
Caribou 指一种生活在寒带/亚寒带的大型鹿科动物,中文常译为: 北美驯鹿 / 北美驯鹿(野生型) / 卡里布驯鹿(不同资料翻译略有差异)
它和 “reindeer(驯鹿)” 在生物学上通常是 同一种动物(同一物种:Rangifer tarandus),只是不同地区习惯叫法不同。
2)Caribou 和 Reindeer 的区别(名字上的区别)
- Caribou:多用于 北美语境(加拿大、阿拉斯加等),常指 野生种群
- Reindeer:多用于 欧洲/亚洲语境,也常指 被人类驯养、放牧的驯鹿
简单记忆:
- 北美野外常叫 caribou
- 欧亚与驯养常叫 reindeer
3)典型特征(为什么它很“特别”)
- 雌雄都有角:很多鹿只有雄鹿长角,但驯鹿/Caribou 的 雌性也常长角。
- 蹄子宽大:像“雪鞋”一样,适合走雪地和沼泽。
- 耐寒结构:毛厚、保温强,适应极冷环境。
- 迁徙行为:许多种群会进行 大规模季节性迁徙。
4)语法:单复数
英语里 caribou 常同时当 单数和复数:
- one caribou(一只)
- three caribou(三只)
也可能看到 caribous,但更常见还是 caribou。
5)单独出现 “Caribou” 可能表示什么
如果只出现一个词 Caribou(没有句子),它很可能是:
- 标题/小标题(这一段讲“驯鹿/北美驯鹿”)
- 图片说明(图下写 Caribou)
- 清单标签(野生动物列表里的一项)
- 象征意义(北境、荒野、迁徙、坚韧等意象)
6)发音
大致读作:“开-ri-bu”(CARE-ih-boo)
If you paste the sentence or paragraph around “Caribou”, I can explain the exact meaning, including whether it’s literal (the animal) or symbolic/metaphorical in that context, in both English and Chinese.
-
-
simonwillison.net simonwillison.net
-
There’s a new kind of coding I call “vibe coding”, where you fully give in to the vibes, embrace exponentials, and forget that the code even exists. It’s possible because the LLMs (e.g. Cursor Composer w Sonnet) are getting too good. Also I just talk to Composer with SuperWhisper so I barely even touch the keyboard. I ask for the dumbest things like “decrease the padding on the sidebar by half” because I’m too lazy to find it. I “Accept All” always, I don’t read the diffs anymore. When I get error messages I just copy paste them in with no comment, usually that fixes it. The code grows beyond my usual comprehension, I’d have to really read through it for a while. Sometimes the LLMs can’t fix a bug so I just work around it or ask for random changes until it goes away. It’s not too bad for throwaway weekend projects, but still quite amusing. I’m building a project or webapp, but it’s not really coding—I just see stuff, say stuff, run stuff, and copy paste stuff, and it mostly works.
vibecoding original description by Andrej Karpathy
Quickly distorted to mean any code created w llm assistance. Note: [[Martijn Aslander p]] follows this dev quite closely (dictation, accept always, it mostly works)
-
-
www.pewresearch.org www.pewresearch.org
-
Pew Research Center has been studying online harassment for several years now. A new report on Americans’ experiences with and attitudes toward online harassment finds that 41% of U.S. adults have personally experienced some form of online harassment – and the severity of the harassment has increased since we last studied it in 2017. We spoke with Emily Vogels, a research associate at the Center focusing on internet and technology research, about the new findings. The interview has been edited for clarity and condensed. One of the big takeaways from this report – and, to me, the biggest surprise – is that, while the overall number of people facing online harassment seems to be more or less stable, the nature of the harassment has changed over time. What are some of the most significant ways in which online harassment has worsened since we first started studying it? Emily Vogels, research associate at Pew Research Center While the overall number of those facing at least one of the six problems we ask about hasn’t changed, this survey finds that the level of harassment is increasing in two key ways: People are more likely to have encountered multiple forms of harassment online, and severe encounters have become more common. When the Center began studying online harassment in 2014, we found that 35% of American adults had experienced it. That grew to 41% in 2017 and remains the same in the new survey. But the shares who have ever experienced more severe forms of harassment – such as physical threats, stalking, sexual harassment or sustained harassment – or multiple forms of harassing behaviors online have both risen substantially in the past three years. This is not the pattern we saw in prior surveys. There has been a markedly steeper rise in these measures since 2017, compared with the change between our 2014 and 2017 studies. The shares who have ever experienced more severe forms of harassment or multiple forms of harassing behaviors online have both risen substantially in the past three years. Also, when we ask people about their most recent harassment experience, they’re more likely than in the past to include these more severe behaviors and involve multiple forms of harassment. And as of 2020, 41% of online harassment targets say their most recent experience spanned multiple locations online – for example, a person being harassed on social media and by text message. Does this suggest that online harassment is, to some extent, becoming “normalized”? It is commonplace. Roughly four-in-ten American adults say they’ve personally experienced harassment online. These numbers are more staggering when we look at adults under 30 – 64% of them say they’ve faced such issues online and 48% say they’ve experienced at least one of the more severe types of harassment. In addition, previous work by the Center found that a majority of adults overall have witnessed others being harassed online. Even when online harassment hasn’t been the focus of our research, we have seen this online incivility play a role in people’s perceptions and experiences of other online phenomena, such as online dating, political discussions on social media and social media in general. The Center’s past research on harassment has shown there are some demographic differences in the kinds of problems people face online. What did this survey show in particular about men, women and harassment? Men are slightly more likely than women to encounter at least one of the six types of online harassment we asked about, but there are notable differences in the types of harassment they encounter. Men are more likely than women to be called an offensive name or be physically threatened. Women are about three times as likely as men to face sexual harassment online, and younger women are even more likely to experience this type of abuse. Another difference in the new survey is that sexual harassment of women has doubled in the past three years, while the rate of sexual harassment among men is largely the same as in 2017. Women who have been the target of online harassment also report finding their most recent harassment experiences to be more upsetting than their male counterparts. There are also differences in where men and women encountered harassment online in their most recent experience. Social media sites are the most common location regardless of gender, but a larger share of women who have been harassed say their most recent incident was on social media, compared with men who have been targeted. Men targeted in online harassment are more likely than women to have been harassed while online gaming or while using an online forum or discussion site. Beyond personal experiences, men and women express different attitudes about online harassment, with women more likely to say it’s a major problem. And prior Center work finds that a greater share of women than men value people feeling safe online over people being able to speak their minds freely. When it comes to how to address online harassment, women are more optimistic than men about a variety of potential solutions, including criminal charges for social media users who harass others online, temporary or permanent bans for users who harass others, and social media companies proactively deleting bullying or harassing posts. Interesting. To what extent do those gender differences in harassment experiences reflect differences in men’s and women’s online activities? Men are more likely to report they had these types of experiences in online forums or gaming platforms. Is that because more men than women use such platforms? It’s a bit complicated. Prior work from the Center suggests there are modest gender differences in gaming, with men being more likely than women to at least sometimes play video games. But this study didn’t ask if people played games online, so we can’t say whether the gender differences in harassment incidents tied to gaming hold when looking at just online gamers. It’s worth keeping in mind that the data on where people were harassed online is for people’s most recent incident, not every incident these folks may have encountered in the past. Prior Center findings show people may stop engaging in an activity – for example, withdrawing from a platform or deleting a social media account – if they encounter harassment. Similarly, do the age differences in those who say they have experienced harassment reflect how many, and how frequently, people of different ages are online? In other words, does the fact that far more adults under 30 report experiencing online harassment reflect younger people spending much more of their lives online than older folks? We don’t quite have enough evidence to make this causal connection, but the broad patterns are pretty clear. This survey found that adults under 30 consistently experience each of the six forms of harassment we asked about at higher rates than any other age group. The Center’s previous work does show that younger adults are more likely to use the internet and to use it almost constantly. Our research on teens in 2018 found that greater exposure to the internet puts people at a higher likelihood of encountering harassment at some point online. It’s worth noting, though, that non-internet users were not asked about their possible experiences with online harassment. So, if people stopped using the internet sometime after they were harassed online, our data wouldn’t capture their earlier harassment experience. The survey finds that 75% of targets of online harassment say their most recent experience was on social media. Has this been true since the Center began researching online harassment? Do people feel social media companies have done enough to discourage this behavior? Fully 79% of Americans think social media companies are doing an only fair to poor job when it comes to addressing online harassment or bullying. The share of online harassment targets who say their most recent harassing encounter took place on social media is growing – up 17 percentage points since 2017. The Center’s prior work reveals a variety of negative opinions Americans hold about social media companies, and when it comes to Americans’ views of how these companies handle online harassment, the pattern of criticism continues. Fully 79% of Americans think social media companies are doing an only fair to poor job when it comes to addressing online harassment or bullying on their platforms. Based on previous Center findings, American teens hold similarly negative views of social media companies’ ability to address these issues. Many Americans suggest that permanent bans for users who harass others and required identity disclosure to use these platforms would be very effective ways to combat harassment on social media. To what extent do you think that the fact 2020 was an election year accounts for the increase in the number of people who say they were harassed because of their political views? Politics was already a heated issue long before this election. According to other research from the Center, partisan antipathy has been growing for years. Americans increasingly say they find they have less in common politically with people with whom they disagree, and they see political discussions online as less respectful, less civil and angrier than political discussions in other places. There are also some striking demographic differences among those who say they’ve been harassed for their politics. Online harassment targets who are White or male – 56% and 57% of each – are particularly likely to think their harassment was a result of their political views. This is especially true for White men who say they’ve been targeted, at 61%. Other groups commonly point to other aspects of their identity as the reason they faced harassment online. For example, roughly half or more Black or Hispanic online harassment targets – 54% and 47% respectively – identify their race or ethnicity as a reason they were harassed, while only 17% of their White counterparts say the same. Bear in mind that politics isn’t the only perceived reason for harassment being on the rise. Over the past several years, rising shares of online harassment targets have said they think they were harassed because of their gender, race, ethnicity, religion or sexual orientation.
The government reports highlight that cyberbullying is widespread and often chronic, affecting many youth for long periods.
-
- Dec 2025
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the original reviews
Public Reviews:
Reviewer #1 (Public review):
Domínguez-Rodrigo and colleagues make a moderately convincing case for habitual elephant butchery by Early Pleistocene hominins at Olduvai Gorge (Tanzania), ca. 1.8-1.7 million years ago. They present this at the site scale (the EAK locality, which they excavated), as well as across the penecontemporaneous landscape, analyzing a series of findspots that contain stone tools and large-mammal bones. The latter are primarily elephants, but giraffids and bovids were also butchered in a few localities. The authors claim that this is the earliest well-documented evidence for elephant butchery; doing so requires debunking other purported cases of elephant butchery in the literature, or in one case, reinterpreting elephant bone manipulation as being nutritional (fracturing to obtain marrow) rather than technological (to make bone tools). The authors' critical discussion of these cases may not be consensual, but it surely advances the scientific discourse. The authors conclude by suggesting that an evolutionary threshold was achieved at ca. 1.8 ma, whereby regular elephant consumption rich in fats and perhaps food surplus, more advanced extractive technology (the Acheulian toolkit), and larger human group size had coincided.
The fieldwork and spatial statistics methods are presented in detail and are solid and helpful, especially the excellent description (all too rare in zooarchaeology papers) of bone conservation and preservation procedures. However, the methods of the zooarchaeological and taphonomic analysis - the core of the study - are peculiarly missing. Some of these are explained along the manuscript, but not in a standard Methods paragraph with suitable references and an explicit account of how the authors recorded bone-surface modifications and the mode of bone fragmentation. This seems more of a technical omission that can be easily fixed than a true shortcoming of the study. The results are detailed and clearly presented.
By and large, the authors achieved their aims, showcasing recurring elephant butchery in 1.8-1.7 million-year-old archaeological contexts. Nevertheless, some ambiguity surrounds the evolutionary significance part. The authors emphasize the temporal and spatial correlation of (1) elephant butchery, (2) Acheulian toolkits, and (3) larger sites, but do not actually discuss how these elements may be causally related. Is it not possible that larger group size or the adoption of Acheulian technology have nothing to do with megafaunal exploitation? Alternative hypotheses exist, and at least, the authors should try to defend the causation, not just put forward the correlation. The only exception is briefly mentioning food surplus as a "significant advantage", but how exactly, in the absence of food-preservation technologies? Moreover, in a landscape full of aggressive scavengers, such excess carcass parts may become a death trap for hominins, not an advantage. I do think that demonstrating habitual butchery bears very significant implications for human evolution, but more effort should be invested in explaining how this might have worked.
Overall, this is an interesting manuscript of broad interest that presents original data and interpretations from the Early Pleistocene archaeology of Olduvai Gorge. These observations and the authors' critical review of previously published evidence are an important contribution that will form the basis for building models of Early Pleistocene hominin adaptation.
This is a good example of the advantages of the eLife reviewing process. It has become much too common, among traditional peer-reviewing journals, to reject articles when there is no coincident agreement in the reviews, regardless of the heuristics (i.e., empirically-supported weight) of the arguments on both reviewers. Reviewers 1 and 2 provide contrasting evaluations, and the eLife dialogue between authors and reviewers enable us to address their comments differentially. Reviewer 1 (R1), whose evaluation is overall positive, remarks that the methods of the zooarchaeological and taphonomic analysis are missing. We have added them now in the revised version of our manuscript. R1 also remarks that our work highlights correlation of events, but not necessarily causation. We did not establish causation because such interpretations bear a considerable amount of speculation (and they might have fostered further criticism by R2); however, in the revised version, we expanded our discussion of these issues substantially. Establishing causation among the events described is impossible, but we certainly provide arguments to link them.
Reviewer #2 (Public review):
The authors argue that the Emiliano Aguirre Korongo (EAK) assemblage from the base of Bed II at Olduvai Gorge shows systematic exploitation of elephants by hominins about 1.78 million years ago. They describe it as the earliest clear case of proboscidean butchery at Olduvai and link it to a larger behavioral shift from the Oldowan to the Acheulean.
The paper includes detailed faunal and spatial data. The excavation and mapping methods appear to be careful, and the figures and tables effectively document the assemblage. The data presentation is strong, but the behavioral interpretation is not supported by the evidence.
The claim for butchery is based mainly on the presence of green-bone fractures and the proximity of bones and stone artifacts. These observations do not prove human activity. Fractures of this kind can form naturally when bones break while still fresh, and spatial overlap can result from post-depositional processes. The studies cited to support these points, including work by Haynes and colleagues, explain that such traces alone are not diagnostic of butchery, but this paper presents them as if they were.
The spatial analyses are technically correct, but their interpretation extends beyond what they can demonstrate. Clustering indicates proximity, not behavior. The claim that statistical results demonstrate a functional link between bones and artifacts is not justified. Other studies that use these methods combine them with direct modification evidence, which is lacking in this case.
The discussion treats different bodies of evidence unevenly. Well-documented cut-marked specimens from Nyayanga and other sites are described as uncertain, while less direct evidence at EAK is treated as decisive. This selective approach weakens the argument and creates inconsistency in how evidence is judged.
The broader evolutionary conclusions are not supported by the data. The paper presents EAK as marking the start of systematic megafaunal exploitation, but the evidence does not show this. The assemblage is described well, but the behavioral and evolutionary interpretations extend far beyond what can be demonstrated.
We disagree with the arguments provided by Reviewer 2 (R2). The arguments are based on two issues: bone breakage and spatial association. We will treat both separately here.
Bone breakage
R2 argues that:
“The claim for butchery is based mainly on the presence of green-bone fractures and the proximity of bones and stone artifacts. These observations do not prove human activity. Fractures of this kind can form naturally when bones break while still fresh, and spatial overlap can result from post-depositional processes. The studies cited to support these points, including work by Haynes and colleagues, explain that such traces alone are not diagnostic of butchery, but this paper presents them as if they were.”
In our manuscript, we argued that green-breakage provides an equally good (or even better) taphonomic evidence of butchery if documented following clear taphonomic indicators. Not all green breaks are equal and not all “cut marks” are unambiguously identifiable as such. First, “natural” elephant long limb breaks have been documented only in pre/peri-mortem stages when an elephant breaks a leg. As a matter of fact, they have only been reported in publication on femora, the thinnest long bone (Haynes et al., 2021). Unfortunately, they have been studied many months after the death of the individuals, and the published diagnosis is made under the assumption that no other process intervened in the modification of those bones during this vast time span. Most of the breaks resulting from pre-mortem fractures produce long smooth, oblique/helical outlines. Occasionally, some flake scarring may occur on the cortical surface. This has been documented as uneven, small-sized, spaced, and we are not sure if it resulted from rubbing of broken fragments while the animal was alive and attempting to walk or some may have resulted from dessication of the bone after one year. When looking at them in detail, such breaks contain sometimes step-microfractures and angular (butterfly-like) outlines. Sometimes, they may be accompanied by pseudo-notches, which are distinct and not comparable to the deep notches that hammerstone breaking generates on the same types of bones. Commonly, the edges of the breaks show some polishing, probably from separate break planes rubbing against each other. It should be emphasized that the experimental work on hammerstone breaking documented by Haynes et al. (2021) is based on bone fracture properties of bones that are no longer completely green. The cracking documented in their hammerstone experimentation, with very irregular outlines differs from the cracking that we are documented in butchery of recently dead elephants.
All this contrasts with the overlapping notches and flake scars (mostly occurring on the medullary side of the bone), both of them bigger in size, with clear smooth, spiral and longitudinal trajectories, with a more intensive modification on the medullary surface, and with sharp break edges resulting from hammerstone breaking of the green bone. No “natural” break has been documented replicating the same morphologies displayed in the Supplementary File to our paper. We display specimens with inflection points, hackle marks on the breaks, overlapping scarring on the medullary surface, with several specimens displaying percussion marks and pitting (also most likely percussion marks). Most importantly, we document this patterned modification on elements other than femora, for which no example has been documented of purported morphological equifinality caused by pre-mortem “natural” breaking. In contrast, such morphologies are documented in hammerstone-broken completely green bones (work in progress). We cited the works of Haynes to support this, because they do not show otherwise. As a matter of fact, Haynes himself had the courtesy of making a thorough reading of our manuscript and did not encounter any contradiction with his work.
Spatial association
R2 argues in this regard:
“The spatial analyses are technically correct, but their interpretation extends beyond what they can demonstrate. Clustering indicates proximity, not behavior. The claim that statistical results demonstrate a functional link between bones and artifacts is not justified. Other studies that use these methods combine them with direct modification evidence, which is lacking in this case.”
We should emphasize that there is some confusion in the use and interpretation of clustering by R2 when applied to EAK. R2 appears to interpret clustering as the typical naked-eye perception of the spatial association of different items. In contrast, we rely on the statistical concept of clustering, more specifically on spatial interdependence or covariance, which is different. Items may appear visually clustered but still be statistically independent. This could, for example, result from two independent depositional episodes that happen to overlap spatially. In such cases, the item-to-item relationship does not necessarily show any spatial interdependence between classes other than simple clustering (i.e., spatial coincidence in intensity).
Spatial statistical interdependence, on the other hand, reflects a spatial relationship or co-dependence between different items. This goes beyond the mere fact that classes appear clustered: items between classes may show specific spatial relationships — they may avoid each other or occupy distinct positions in space (regular co-dependence), or they may interact within the same spatial area (clustering co-dependence). Our tests indicate the latter for EAK.
Such patterns are difficult to explain when depositional events are unrelated, since the probability that two independent events would generate identical spatial patterns in the same loci is very low. They are also difficult to reconcile when post-depositional processes intervene and resediment part of the assemblage (Domínguez-Rodrigo et al. 2018).
Finally, R2 concludes:
“The discussion treats different bodies of evidence unevenly. Well-documented cut-marked specimens from Nyayanga and other sites are described as uncertain, while less direct evidence at EAK is treated as decisive. This selective approach weakens the argument and creates inconsistency in how evidence is judged.”
The Nyayanga hippo remains bearing modifications have not been well-documented cut marks. Neither R2 nor we can differentiate those marks from those inflicted by natural abrasive processes in coarse-grained sedimentary contexts, where the carcasses are found. The fact that the observable microscopic features (through low-quality photographs as appear in the original publication) differ between the cut marks documented on smaller animals and those inferred for the hippo remains makes them even more ambiguous. Nowhere in our manuscript do we treat the EAK evidence (or any other evidence) as decisive, but as the most likely given the methods used and the results reported.
References
Haynes G, Krasinski K, Wojtal P. 2021. A Study of Fractured Proboscidean Bones in Recent and Fossil Assemblages. Journal of Archaeological Method and Theory 28:956–1025.
Domínguez-Rodrigo, M., Cobo-Sánchez, L., yravedra, J., Uribelarrea, D., Arriaza, C., Organista, E., Baquedano, E. 2018. Fluvial spatial taphonomy: a new method for the study of post-depositional processes. Archaeological and Anthropological Sciences 10: 1769-1789.
Recommendations for authors:
Reviewer #1 (Recommendations for the authors):
I have several recommendations that, in my opinion, could enhance the communication of this study to the readers. The first point is the only crucial one.
(1) A detailed zooarchaeological methods section must be added, with explanations (or references to them) of precisely how the authors defined and recorded bone-surface modifications and mode of bone fragmentation.
This appears in the revised version of the manuscript in the form of a new sub-section within the Methods section.
(2) The title could be improved to better represent the contents of the paper. It contains two parts: the earliest evidence for elephant butchery (that's ok), and revealing the evolutionary impact of megafaunal exploitation. The latter point is not actually revealed in the manuscript, just alluded to here and there (see also below).
We have elaborated on this in the revised version, linking megafaunal exploitation and anatomical changes (which appear discussed in much more detail in the references indicated).
(3) The abstract does not make it clear whether the authors think that the megafaunal adaptation strongly correlates with the Acheulian technocomplex. It seems that they do, so please make this point apparent in the abstract.
From a functional point of view, we document the correlation, but do not believe in the causation, since most butchering tools around these megafaunal carcasses are typologically non Acheulian. We have indicated so in the abstract.
(4) Please define what you mean by "megafauna". How large should an animal be to be considered as megafauna in this particular context?
We have added this definition: we identify as “megafauna” those animals heavier than 800 kg.
(5) In the literature survey, consider also this Middle Pleistocene case-study of elephant butchery, including a probable bone tool: Rabinovich, R., Ackermann, O., Aladjem, E., Barkai, R., Biton, R., Milevski, I., Solodenko, N., and Marder, O., 2012. Elephants at the middle Pleistocene Acheulian open-air site of Revadim Quarry, Israel. Quaternary International, 276, pp.183-197.
Added to the revised version
(6) The paragraph in lines 123-160 is unclear. Do the authors argue that the lack of evidence for processing elephant carcasses for marrow and grease is universal? They bring forth a single example of a much later (MIS 5) site in Germany. Then, the authors state the huge importance of fats for foragers (when? Where? Surely not in all latitudes and ecosystems). This left me confused - what exactly are you trying to claim here?
We have explained this a little more in the revised text. What we pointed out was that most prehistoric (and modern) elephant butchery sites leave grease-containing long bones intact. Evidence of anthropogenic breakage of these elements is rather limited. The most probably reason is the overabundance of meat and fat from the rest of the carcass and the time-consuming effort needed to access the medullary cavity of elephant long bones.
(7) The paragraph in lines 174-187 disrupts the flow of the text, contains previously mentioned information, ends with an unclear sentence, and could be cut.
(8) Results: please provide the MNI for the EAK site (presumably 1, but this is never mentioned).
Done in the revised version.
(9) Lines 292 - 295: The authors found no traces of carnivoran activity (carnivoran remains, coprolites, or gnawing marks on the elephant bones), yet they attribute the absence of some non-dense skeletal elements to carnivore ravaging. I cannot understand this rationale, given that other density-mediated processes could have deleted the missing bones and epiphysis.
This interpretation stems from our observations of several elephant carcasses in the Okavango delta in Botswana. Those that were monitored showed deletion of remains (i.e., disappearance of certain bones, like feet) without necessarily imprinting damage on the rest of the carcass. Carnivore intervention in an elephant death site can result in deletion of a few remains without much damage (if any), or if hyena clans access the carcass, much more conspicuous damage can be documented. There is a whole range of carnivore signatures in between. We are currently working on our study of several elephant carcasses subjected to these highly variable degrees of carnivore impact.
(10) Lines 412 - 422: "The clustering of the elephant (and hippopotamus) carcasses in the areas containing the highest densities of landscape surface artifacts is suggestive of a hominin agency in at least part of their consumption and modification." - how so? It could equally suggest that both hominins and elephants were drawn to the same lush environments.
We agree. Both hominins and megafauna must have been drawn to the same ecological loci for interaction to emerge. However, the fact that the highest density clusters of artifacts coincide with the highest density of carcasses “showing evidence of having been broken”, is suggestive of hominin use and consumption.
(11) Discussion: I suggest starting the Discussion with a concise appraisal of the lines of evidence detailed in the Results and their interpretation, and only then, the critical reassessment of other studies. Similarly, a new topic starts in line 508, but without any subheading or an introductory sentence that could assist the readers.
We added the introductory lines of the former Conclusion section to the revised Discussion section, as suggested by R1.
(12) Line 607: Neumark-Nord are Late Pleistocene sites (MIS 5), not Middle Pleistocene.
Corrected.
(13) Regarding the ambiguity in how megafaunal exploitation may be causally related to the other features of the early Acheulian, the authors can develop the discussion. Alternatively, they should explicitly state that correlation is not causation, and that the present study adds the megafaunal exploitation element to be considered in future discussion of the shifts in lifestyles 1.8 million years ago.
We have done so.
Reviewer #2 (Recommendations for the authors):
The following detailed comments are provided to help clarify arguments, ensure accurate representation of cited literature, and strengthen the logical and methodological framing of the paper. Line numbers refer to the version provided for review.
(1) Line 55: Such concurrency (sometimes in conjunction with other variables)
The term "other variables" is very vague. I would suggest expanding on this or taking it out altogether.
(2) Line 146: Megafaunal long bone green breakage (linked to continuous spiral fractures on thick cortical bone) is probably a less ambiguous trace of butchery than "cut marks", since many of the latter could be equifinal and harder to identify, especially in contexts of high abrasion and trampling (Haynes et al., 2021, 2020).
This reasoning is not supported by the evidence or the cited sources. Green-bone spiral fractures only show that a bone broke while it was fresh and do not reveal who or what caused it. Carnivore feeding, trampling, and natural sediment pressure can all create the same patterns, so these fractures are not clearer evidence of butchery than cut marks. Cut marks, when they are preserved and morphologically clear, remain the most reliable indicator of human activity. The Haynes papers actually show the opposite of what is claimed here. They warn that spiral fractures and surface marks can form naturally and that fracture patterns alone cannot be used to infer butchery. This section should be revised to reflect what those studies actually demonstrate.
The reasoning referred to in line 146 is further explained below in the original text as follows:
“Despite the occurrence of green fractures on naturally-broken bones, such as those trampled by elephants (Haynes et al., 2020), those occurring through traumatic fracturing or gnawed by carnivores (Haynes and Hutson, 2020), these fail to reproduce the elongated, extensive, or helicoidal spiral fractures (uninterrupted by stepped sections), accompanied by the overlapping conchoidal scars (both cortical and medullary), the reflected scarring, the inflection points, or the impact hackled break surfaces and flakes typical of dynamic percussive breakage. Evidence of this type of green breakage had not been documented earlier for the Early Pleistocene proboscidean or hippopotamid carcasses, beyond the documentation of flaked bone with the purpose of elaboration of bone tools (Backwell and d’Errico, 2004; Pante et al., 2020; Sano et al., 2020).”
The problem in the way that R2 uses Haynes et al.´s works is that R2 uses features separately. Natural breaks occurring while the bone is green can generate spiral smooth breaks, for example, but it is not the presence of a single feature that invalidates the diagnosis of agency or that is taphonomically relevant, but the concurrence of several of them. The best example of a naturally (pre-mortem) broken bone was published by Haynes et al.
The natural break shows helical fractures, subjugated to linear (angular) fracture outlines. Notice how the crack displays a zig-zag. The break is smooth but most damage occurs on the cortical surface, with flaking adjacent to the break and step micro-fracturing on the edges. The cortical scarring is discontinuous (almost marginal) and very small, almost limited to the very edge of the break. No modification occurs on the medullary surface. No extensive conchoidal fractures are documented, and certainly none inside the medullary surface of the break.
Compare with Figure S8, S10, S17 and S34 (all specimens are shown in their medullary surface):
In these examples, we see clearly modified medullary surfaces with multiple green breaks and large-sized step fractures, accompanied in some examples by hackle marks. Some show large overlapping scars (of substantially bigger size than those documented in the natural break image). Not a single example of naturally-broken bones has been documented displaying these morphologies simultaneously. It is the comprehensive analysis of the co-occurrence of these features and not their marginal and isolated occurrence in naturally-broken bones that make a difference in the attribution of agency. Likewise, no example of naturally-broken bone has been published that could mimic any of the two green-broken bones documented at EAK. In contrast, we do have bones from our on-going experimentation with green elephant carcasses that jointly reproduce these features. See also Figure 6 of the article to find another example without any modern referent in the naturally-broken bones documented.
We should emphasize that R2 is inaccurately portraying what Haynes et al.´s results really document. Contrary to R2´s assertion, trampling does not reproduce any of the examples shown above. Neither do carnivores. It should be stressed that Haynes & Harrod only document similar overlapping scarring on the medullary surface of bones, when using much smaller animals. In all the carnivore damage repertoire that they document for elephants, durophagous spotted hyenas can only inflict furrowing on the ends of the biggest long bones, especially if they are adults. Long bone midshafts remain inaccessible to them. The mid-shaft portions of bones that we document in our Supplementary File and at EAK cannot be the result of hyena (or carnivore damage) for this reason, and also because their intense gnawing on elephant bones leaves tooth marking on most of the elements that they modify, being absent in our sample.
(3) Line 176: other than hominins accessed them in different taphonomically-defined stages- stages - the "Stages" is repeated twice
Defined in the revised version
(4) Line 174: Regardless of the type of butchery evidence - and with the taphonomic caveat that no unambiguous evidence exists to confirm that megafaunal carcasses were hunted or scavenged other than hominins accessed them in different taphonomically-defined stages- stages - the principal reasons for exploring megafaunal consumption in early human evolution is its origin, its episodic or temporally-patterned occurrence, its impact on hominin adaptation to certain landscapes, and its reflection on hominin group size and site functionality.
This sentence is confusing and needs to be rewritten for clarity. It tries to combine too many ideas at once, and the phrasing makes it hard to tell what the main point is. The taphonomic caveat in the middle interrupts the sentence and obscures the argument. It should be broken into separate, clearer statements that distinguish what evidence exists, what remains uncertain, and what the broader goals of the discussion are.
We believe the ideas are displayed clearly
(5) Line 179: landscapes, and its reflection on hominin group size and site functionality. If hominins actively sought the exploitation of megafauna, especially if targeting early stages of carcass consumption, the recovery of an apparent surplus of resources reflects a substantially different behavior from the small-group/small-site pattern documented at several earlier Oldowan anthropogenic sites (Domínguez-Rodrigo et al., 2019) -or some modern foragers, like the Hadza, who only exploit megafaunal carcasses very sporadically, mostly upon opportunistic encounters (Marlowe, 2010; O'Connell et al., 1992; Wood, 2010; Wood and Marlowe, 2013).
This sentence makes a reasonable point, but is written in a confusing way. The idea that early, deliberate access to megafauna would represent a different behavioral pattern from smaller Oldowan or modern foraging contexts is valid, but the sentence is awkward and hard to follow. It should be rephrased to make the logic clearer and more direct.
We believe the ideas are displayed clearly
(6) Line 186: When the process started of becoming megafaunal commensal started has major implications for human evolution.
This sentence is awkward and needs to be rewritten for clarity. The phrasing "when the process started of becoming megafaunal commensal started" is confusing and grammatically incorrect. It could be revised to something like "Determining when hominins first began to interact regularly with megafauna has major implications for human evolution," or another version that clearly identifies the process being discussed.
Modified in the revised version
(7) Line189: The multiple taphonomic biases intervening in the palimpsestic nature of most of these butchery sites often prevent the detection of the causal traces linking megafaunal carcasses and hominins. Functional links have commonly been assumed through the spatial concurrence of tools and carcass remains; however, this perception may be utterly unjustified as we argued above. Functional association of both archaeological elements can more securely be detected through objective spatial statistical methods. This has been argued to be foundational for heuristic interpretations of proboscidean butchery sites (Giusti, 2021). Such an approach removes ambiguity and solidifies spatial functional association, as demonstrated at sites like Marathousa 1 (Konidaris et al., 2018) or TK Sivatherium (Panera et al., 2019). This method will play a major role in the present study.
This section overstates what spatial analysis can demonstrate and misrepresents the cited studies. The works by Giusti (2021), Konidaris et al. (2018), and Panera et al. (2019) do use spatial statistics to examine relationships between artifacts and faunal remains, but they explicitly caution that spatial overlap alone does not prove functional or behavioral association. These studies argue that clustering can support such interpretations only when combined with detailed taphonomic and stratigraphic evidence. None of them claims that spatial analysis "removes ambiguity" or "solidifies" functional links. The text should be revised to reflect the more qualified conclusions of those papers and to avoid implying that spatial statistics can establish behavioral causation on their own.
We disagree. Both works (Giusti and Panera) use spatial statistical tools to create an inferential basis reinforcing a functional association of lithics and bones. In both cases, the anthropogenic agency inferred is based on that. We should stress that this only provides a basis for argumentation, not a definitive causation. Again, those analyses show much more than just apparent visual clustering.
(8) Line 200: Here, we present the discovery of a new elephant butchery site (Emiliano Aguirre Korongo, EAK), dated to 1.78 Ma, from the base of Bed II at Olduvai Gorge. It is the oldest unambiguous proboscidean butchery site at Olduvai.
It is fine to state the main finding in the introduction, but the phrasing here is too strong. Calling EAK "the oldest unambiguous proboscidean butchery site" asserts certainty before the evidence is presented. The claim should be stated more cautiously, for example, "a new site that provides early evidence for proboscidean butchery," so that the language reflects the strength of the data rather than pre-judging it.
We understand the caution by R2, but in this case, EAK is the oldest taphonomically-supported evidence of elephant butchery at Olduvai (see discussion about FLK North in the text). Whether this is declared at the beginning or the end of the text is irrelevant.
(9) Line 224: The drying that characterizes Bed II had not yet taken place during this moment.
This sentence reads like a literal translation. It should be rewritten for clarity.
Modified in the revised version
(10) Line 233: During the recent Holocene, the EAK site was affected by a small landslide which displaced the...
This section contains far more geological detail than is needed for the argument. The reader only needs to know that the site block was displaced by a small Holocene landslide but retains its stratigraphic integrity. The extended discussion of regional faults, seismicity, and slope processes goes well beyond what is necessary for context and distracts from the main focus of the paper.
We disagree. The geological information is what is most commonly missing from most archaeological reports. Here, it is relevant because of the atypical process and because it has been documented only twice with elephant butchery sites. Explaining the dynamic geological process that shaped the site helps to understand its spatial properties.
(11) Line 264: In June 2022, a partial elephant carcass was found at EAK on a fragmented stratigraphic block...
This section reads like field notes rather than a formal site description. Most of the details about the discovery sequence, trench setup, and excavation process are unnecessary for the main text. Only the basic contextual information about the find location, stratigraphic position, and anatomical composition is needed. The rest could be condensed or moved to the methods or supplementary material.
We disagree. See reply above.
(12) Line 291: hominins or other carnivores. Ongoing restoration work will provide an accurate estimate of well-preserved and modified fractions of the assemblage.
This sentence is unclear and needs to specify what kind of restoration work is being done and what is meant by well-preserved and modified fractions. It is not clear whether modified refers to surface marks, diagenetic alteration, or something else. If the bones are still being cleaned or prepared, the analysis is incomplete, and the counts cannot be considered final. If restoration only means conservation or stabilization, that should be stated clearly so the reader understands that it does not affect the results. As written, it is not clear whether the data presented here are preliminary or complete.
We added: For this reason, until restoration is concluded, we cannot produce any asssertion about the presence or absence of bone surface modifications.
(13) Line 294: The tibiae were well preserved, but the epiphyseal portions of the femora were missing, probably removed by carnivores, which would also explain why a large portion of the rib cage and almost all vertebrae are missing.
This explanation is not well supported. The missing elements could be the result of other forms of density-mediated destruction, such as sediment compaction or post-depositional fragmentation, especially since no tooth marks were found. Given the low density of ribs, vertebrae, and femoral epiphyses, these processes are more likely explanations than carnivore removal. The text should acknowledge these alternatives rather than attributing the pattern to carnivore activity without direct evidence.
Sediment compaction and post-depositional can break bones but cannot make them disappear. Our excavation process was careful enough to detect bone if present. Their absence indicates two possibilities: erosion through the years at the front of the excavation or carnivore intervention. Carnivores can take elephant bones without impacting the remaining assemblage (see our reply above to a similar comment).
(14) Line 304: The fact that the carcass was moved while encased in its sedimentary context, along with the close association of stone tools with the elephant bones, is in agreement with the inference that the animal was butchered by hominins. A more objective way to assess this association is through spatial statistical analysis.
The authors state that "the carcass was moved while encased in its sedimentary context, along with the close association of stone tools with the elephant bones, is in agreement with the inference that the animal was butchered by hominins." This does not logically follow. Movement of the block explains why the bones and tools remain together, not how that association was created. The preserved association alone does not demonstrate butchery, especially in the absence of cut marks or other direct evidence of hominin activity.
Again, we are sorry that R2 is completely overlooking the strong signal detected by the spatial statistical analysis. The way that the block moved, it preserved the original association of bones and tools. This statement is meant to clarify that despite the allochthonous nature of the block, the original autochthonous depositional process of both types of archaeological materials has been preserved. The spatial association, as statistically demonstrated, indicates that the functional link is more likely than any other alternative process. The additional fact that nowhere else in that portion of the outcrop do we identify scatters of tools (all appear clustered at a landscape scale with the elephant) adds more support to this interpretation. This would have been further supported by the presence of cut marks, no doubt, but their absence does not indicate lack of functional association, since as Haynes´ works have clearly shown, most bulk defleshing of modern elephant leaves no traces on most bones.
(15) Line 370: This also shows that the functional connection between the elephant bones and the tools has been maintained despite the block post-sedimentary movement.
The spatial analyses appear to have been carried out appropriately, and the interpretations of clustering and segregation are consistent with the reported results. However, the conclusion that the "functional connection" between bones and tools has been maintained goes beyond what spatial correlation alone can demonstrate. These analyses show spatial proximity and scale-dependent clustering but cannot, by themselves, confirm a behavioral or functional link.
R2 is making this comment repeatedly and we have addressed it more than once above. We disagree and we refer to our replies above to sustain it.
(16) Line 412: The clustering of the elephant (and hippopotamus) carcasses in the areas containing the highest densities of landscape surface artifacts is suggestive of a hominin agency in at least part of their consumption and modification. The presence of green broken elephant long bone elements in the area surveyed is only documented within such clusters, both for lower and upper Bed II. This constitutes inverse negative evidence for natural breaks occurring on those carcasses through natural (i.e., non-hominin) pre- and peri-mortem limb breaking (Haynes et al., 2021, 2020; Haynes and Hutson, 2020). In this latter case, it would be expected for green-broken bones to show a more random landscape distribution, and occur in similar frequencies in areas with intense hominin landscape use (as documented in high density artifact deposition) and those with marginal or non-hominin intervention (mostly devoid of anthropogenic lithic remains).
The clustering of green-bone fractures with stone tools is intriguing but should be interpreted cautiously. The Haynes references are misrepresented here. Those studies address both cut marks and green-bone (spiral) fractures, emphasizing that each can arise through non-hominin processes such as trampling, carcass collapse, and sediment loading. They do not treat green fractures as clearer evidence of butchery; in fact, they caution that such breakage patterns can occur naturally and even form clustered distributions in areas of repeated animal activity. The claim that these studies support spiral fractures as unambiguous indicators of hominin activity, or that natural breaks would be randomly distributed, is not accurate.
We would like to emphasize again that the Haynes´references are not misrepresented here. See our extensive reply above. If R2 can provide evidence of natural breakage patterns resulting from pre-mortem limb breaking or post-mortem trampling resulting in all limb bones being affected by these processes and resulting in smooth spiral breaks, accompanied with extensive and overlapping scarring on the medullary surface, in conjunction with the other features described in our replies above, then we would be willing to reconsider. With the evidence reported until now, that does not occur simultaneously on specimens resulting from studies on modern elephant bones.
R2 seems to contradict him(her)self here by saying that Haynes studies show that cut marks are not reliable because they can also be reproduced via trampling. Until this point, R2 had been saying that only cut marks could demonstrate a functional link and support butchery. Haynes´ studies do not deal experimentally with sediment loading.
(17) Line 424: This indicates that from lower Bed II (1.78 Ma) onwards, there is ample documented evidence of anthropogenic agency in the modification of proboscidean bones across the Olduvai paleolandscapes. The discovery of EAK constitutes, in this respect, the oldest evidence thereof at the gorge. The taphonomic evidence of dynamic proboscidean bone breaking across time and space supports, therefore, the inferences made by the spatial statistical analyses of bones and lithics at the site.
This conclusion is overstated. The claim of "ample documented evidence of anthropogenic agency" is too strong, given that the main support comes from indirect indicators like green-bone fractures and spatial clustering rather than clear butchery marks. It would be more accurate to say that the evidence suggests or is consistent with possible hominin involvement. The final sentence also conflates association with causation; spatial and taphonomic data can indicate a relationship, but do not confirm that the carcasses were butchered by hominins.
The evidence is based on spatially clustering (at a landscape scale) of tools and elephant (and other megafaunal taxa) bones, in conjunction with a large amount of green-broken elements. This interpretation, if we compare it against modern referents is supported even stronger. In the past few years, we have been conducting work on modern naturally dead elephant carcasses in Botswana and Zambia, and of the several carcasses that we have seen, we have not identified a single case of long bone shaft breaks like those described by Haynes as natural or like those we describe here as anthropogenic. This probably means that they are highly unlikely or marginal occurrences at a landscape scale. This seems to be supported by Haynes´ work too. Out of the hundreds of elephant carcasses that he has monitored and studied over the years for different works, we have managed to identify only two instances where he described natural pre-mortem breaks. This certainly qualifies as extremely marginal.
Most of the Results section is clearly descriptive, but beginning with "The clustering of the elephant (and hippopotamus) carcasses..." the text shifts from reporting observations to drawing behavioral conclusions. From this point on, it interprets the data as evidence of hominin activity rather than simply describing the patterns. This part would be more appropriate for the Discussion, or should be rewritten in a neutral, descriptive way if it is meant to stay in the Results.
This appears extensively discussed in the Discussion section, but the data presented in the results is also interpreted in that section, following a clear argumental chain.
(18) Line 433: A recent discovery of a couple of hippopotamus partial carcasses at the 3.0-2.6 Ma site of Nyayanga (Kenya), spatially concurrent with stone artifacts, has been argued to be causally linked by the presence of cut marks on some bones (Plummer et al., 2023). The only evidence published thereof is a series of bone surface modifications on a hippo rib and a tibial crest, which we suggest may be the result of byproduct of abiotic abrasive processes; the marks contrast noticeably with the well-defined cut marks found on smaller mammal bones (Plummer et al. ́s 2023: Figure 3C, D) associated with the hippo remains (Plummer et al., 2023).
The authors suggest that the Nyayanga marks could result from abiotic abrasion, but this claim does not engage with the detailed evidence presented by Plummer et al. (2023). Plummer and colleagues documented well-defined, morphologically consistent cut marks and considered the sedimentary context in their interpretation. Raising abrasion as a general possibility without addressing that analysis gives the impression of selective skepticism rather than an evaluation grounded in the published data.
We disagree again on this matter. R2 does not clarify what he/she means by well-defined or morphologically consistent. We provide an alternative interpretation of those marks that fit their morphology and features and that Plummer at al did not successfully exclude. We also emphasize that the interpretation of the Nyayanga marks was made descriptively, without any analytical approach and with a high degree of subjectivity by the researcher. All of this disqualifies the approach as well defined and keeps casting an old look at modern taphonomy. Descriptive taphonomy is a thing of the 1980´s. Today there are a plethora of analytical methods, from multivariate statistics, to geometric morphometrics to AI computer vision (so far the most reliable) which represent how taphonomy (and more specifically, analysis of bone surface modifications) should be conducted in the XXI century. This approaches would reinforce interpretations as preliminarily published by Plummer et al, provided they reject alternative explanations like those that we have provided.
(19) Line 459: It would have been essential to document that the FLK N6 tools associated with the elephant were either on the same depositional surface as the elephant bones and/or on the same vertical position. The ambiguity about the FLK N6 elephant renders EAK the oldest secure proboscidean butchery evidence at Olduvai, and also probably one of the oldest in the early Pleistocene elsewhere in Africa.
The concern about vertical mixing is fair, but the tone makes it sound like the association is definitely not real. It would be more accurate to say that the evidence is ambiguous, not that it should be dismissed altogether.
We have precisely done so. We do not dismiss it, but we cannot take it for anything solid since we excavated the site and show how easily one could make functional associations if forgetting about the third dimension. It is not a secure butchery site. This is what we said and we stick to this statement.
(20) Line 479: In all cases, these wet environments must have been preferred places for water-dependent megafauna, like elephants and hippos, and their overlapping ecological niches are reflected in the spatial co-occurrence of their carcasses. Both types of megafauna show traces of hominin use through either cutmarked or percussed bones, green-broken bones, or both (Supplementary Information).
The environmental part is good, but the behavioral interpretation is too strong. Saying elephants and hippos "must have been" drawn to these areas is too certain, and claiming that both "show traces of hominin use" makes it sound like every carcass was modified. It should be clearer that only some have possible evidence of this.
The sentence only refers to both types of fauna taxonomically. No inference can be drawn therefor that all carcasses are modified.
(21) Line 496: In most green-broken limb bones, we document the presence of a medullary cavity, despite the continuous presence of trabecular bone tissue on its walls.
This sentence is confusing and doesn't seem to add anything meaningful. All limb bones naturally have a medullary cavity lined with trabecular bone, so it's unclear why this is noted as significant. The authors should clarify what they mean here or remove it if it's simply describing normal bone structure.
No. Modern elephant long bones do not have a hollow medullary cavity. All the medullary volume is composed of trabecular tissue. Some elephants in the past had hollow medullary cavities, which probably contained larger amounts of marrow and fat.
(22) Line 518: We are not confident that the artefacts reported by de la Torre et al are indeed tools.
While I generally agree with this statement, the paragraph reads as defensive rather than comparative. It would help if they briefly summarized what de la Torre et al. actually argued before explaining why they disagree.
We devote two full pages of the Discussion section to do so precisely.
(23) Lines 518-574: They are similar to the green-broken specimens that we have reported here...
This part is very detailed but inconsistent. They argue that the T69 marks could come from natural processes, but they use similar evidence (green fractures, overlapping scars) to argue for human activity at EAK. If equifinality applies to one, it applies to both.
We are confused by this misinterpretation. Features like green fractures and overlapping scars (among others) can be used to detect anthropogenic agency in elephant bone breaking; that is, any given specimen can be determined to have been an “artifact” (in the sense of human-created item), but going from there to interpreting an artifact as a tool, there is a large distance. Whereas an artifact (something made by a human) can be created indirectly through several processes (for example, demarrowing a bone resulting in long bone fragments), a tool suggest either intentional manufacture and use or both. That is the difference between de la Torre et al.´s interpretation and ours. We believe that they are showing anthropogenically-made items, but they have provided no proof that they were tools.
(24) Line 576: A final argument used by the authors to justify the intentional artifactual nature of their bone implements is that the bone tools were found in situ within a single stratigraphic horizon securely dated to 1.5 million years ago, indicating systematic production rather than episodic use. This is taphonomically unjustified.
The reasoning here feels uneven in how clustering evidence is used. At EAK, clustering of bones and artifacts is taken as meaningful evidence of hominin activity, but here the same pattern at T69 is treated as a natural by-product of butchery or carnivore activity. If clustering alone cannot distinguish between intentional and incidental association, the authors should clarify why it is interpreted as diagnostic in one case but not in the other.
Again, we are confused by this misinterpretation. It applies to two different scenarios/questions:
a) is there a functional link between tools and bones at EAK and T69? We have statistically demonstrated that at EAK and we think de la Torre et al. is trying to do the same for T69, although using a different method.
b) Are the purported tools at T69 tools? Are those that we report here tools? In this regard there is no evidence for either case and given that several bones from T69 come from animals smaller than elephants, we do not discard that carnivores might have been responsible for those, whereas hominin butchery might have been responsible for the intense long limb breaking at that site. It remains to be seen how many (if any) of those specimens were tools.
(25) Line 600: If such a bone implement was a tool, it would be the oldest bone tool documented to date (>1.7 Ma).
The comparison to prior studies is useful, and the point about missing use-wear traces is well taken. However, the last lines feel speculative. If no clear use evidence has been found, it's premature to suggest that one specimen "would be the oldest bone tool." That claim should be either removed or clearly stated as hypothetical.
It clearly reads as hypothetical.
(26) Line 606: Evidence documents that the oldest systematic anthropogenic exploitation of proboscidean carcasses are documented (at several paleolandscape scales) in the Middle Pleistocene sites of Neumark-Nord (Germany)(Gaudzinski-Windheuser et al., 2023a, 2023b).
This is the first and only mention of Neumark-Nord in the paper, and it appears without any prior discussion or connection to the rest of the study. If this site is being used for comparison or as part of a broader temporal framework, it needs to be introduced and contextualized earlier. As written, it feels out of place and disconnected from the rest of the argument.
This is a Late Pleistocene site and we do not see the need to present it earlier, given that the scope of this work is Early Pleistocene.
(27) Line 608: Evidence of at least episodic access to proboscidean remains goes back in time (see review in Agam and Barkai, 2018; Ben-Dor et al., 2011; Haynes, 2022).
The distinction between "systematic" and "episodic" exploitation is useful, but the authors should clarify what criteria define each. The phrase "episodic access...goes back in time" is vague and could be replaced with a clearer statement summarizing the nature of the earlier evidence.
It is self-explanatory
(28) Line 610: Redundant megafaunal exploitation is well documented at some early Pleistocene sites from Olduvai Gorge (Domínguez-Rodrigo et al., 2014a, 2014b; Organista et al., 2019, 2017, 2016).
The phrase "redundant megafaunal exploitation" needs clarification. "Redundant" is not standard terminology in this context. Does this mean repeated, consistent, or overlapping behaviors? Also, while these same Olduvai sites are mentioned earlier, this phrasing also introduces new interpretive language not used before and implies a broader behavioral generalization than what the data actually show.
Webster: Redundant means repetitive, occurring multiple times.
(29) Line 612: At the very same sites, the stone artifactual assemblages, as well as the site dimensions, are substantially larger than those documented in the Bed I Oldowan sites (Diez-Martín et al., 2024, 2017, 2014, 2009).
The placement and logic of this comparison are unclear. The discussion moves from Middle Pleistocene Neumark-Nord to early Pleistocene Olduvai sites, then to Bed I Oldowan contexts without clearly signaling the temporal or geographic transitions. If the intent is to contrast Acheulean vs. Oldowan site scale or organization, that connection needs to be made explicit. As written, it reads as a disjointed shift rather than a continuation of the argument.
We disagree. Here, we finalize by bringing in some more recent assemblages where hominin agency is not in question.
(30) Line 616: Here, we have reported a significant change in hominin foraging behaviors during Bed I and Bed II times, roughly coinciding with the replacement of Oldowan industries by Acheulian tool kits -although during Bed II, both industries co-existed for a substantial amount of time (Domínguez-Rodrigo et al., 2023; Uribelarrea et al., 2019, 2017).
This section should be restructured for flow. The reference to behavioral change during Bed I-II and the overlap of Oldowan and Acheulean industries is important, but feels buried after a long detour. Consider moving this earlier or rephrasing so the main conclusion (behavioral change across Beds I-II) is clearly stated first, followed by supporting examples.
It is not within the scope of this work and is properly described in the references mentioned.
(31) Line 620: The evidence presented here, together with that documented by de la Torre et al. (2025), represents the most geographically extensive documentation of repeated access to proboscidean and other megafaunal remains at a single fossil locality.
The phrase "most geographically extensive documentation of repeated access" overstates what has been demonstrated. The evidence presented is site-specific and does not justify such a broad superlative. This should be toned down or supported with comparative quantitative data.
We disagree. There is no other example where such an abundant record of green-broken elements from megafauna is documented. Neumark-Nord is more similar because it shows extensive evidence of butchery, but not so much about degreasing.
(32) Line 623: The transition from Oldowan sites, where lithic and archaeofaunal assemblages are typically concentrated within 30-40 m2 clusters, to Acheulean sites that span hundreds or even over 1000 m2 (as in BK), with distinct internal spatial organization and redundancy in space use across multiple archaeological layers spanning meters of stratigraphic sequence (Domínguez-Rodrigo et al., 2014a, 2009b; Organista et al., 2017), reflects significant behavioral and technological shifts.
This sentence about site size and spatial organization repeats earlier claims without adding new insight. If it's meant as a synthesis, it should explicitly say how the spatial expansion relates to changes in behavior or mobility, not just describe the difference.
In the Conclusion section these correlations have been explained in more detail to add some causation.
(33) Line 628: This pattern likely signifies critical innovations in human evolution, coinciding with major anatomical and physiological transformations in early hominins (Dembitzer et al., 2022; Domínguez-Rodrigo et al., 2021, 2012).
The conclusion that this "signifies critical innovations in human evolution" is too sweeping, given the data presented. It introduces physiological and anatomical transformation without connecting it to any evidence in this paper. Either cite the relevant findings or limit the claim to behavioral implications.
The references cited elaboration in extension this. The revised version of the Conclusion section also elaborates on this.
Overall, the conclusions section reads as a loosely connected set of assertions rather than a focused synthesis. It introduces new interpretations and terminology not supported or developed earlier in the paper, and the argument jumps across temporal and geographic scales without clear transitions. The discussion should be restructured to summarize key results, clarify the scope of interpretation, and avoid speculative or overstated claims about evolutionary significance.
We have done so, supported by the references used in addition to extending some of the arguments
(34) Line 639: The systematic excavation of the stratigraphic layers involved a small crew.
This sentence is not necessary.
No comment
(35) Line 643: The orientation and inclination of the artifacts were recorded using a compass and an inclinometer, respectively.
What were these measurements used for (e.g., post-depositional movement analysis, spatial patterning)? A short note on the purpose would make this more meaningful.
Fabric analysis has been added to the revised version.
(36) Line 659: Restoration of the EAK elephant bones
This section could be streamlined and clarified. It includes procedural detail that doesn't contribute to scientific replicability (e.g., the texture of gauze, number of consolidant applications), while omitting some key information (such as how restoration may have affected analytical results). It also contains interpretive comments ("most of the assemblage has been successfully studied") that don't belong in Methods.
No comment
(37) Line 689: In the field laboratory, cleaning of the bone remains was carried out, along with adhesion of fragments and their consolidation when necessary.
Clarify whether cleaning or adhesion treatments might obscure or alter bone surface modifications, as this has analytical implications.
These protocols do not impact bone like that anymore.
(38) Line 711: (b) Percussion Tools - Includes hammerstones or cobbles exhibiting diagnostic battering, pitting, and/or impact scars consistent with percussive activities.
Define how diagnostic features (battering, pitting) were identified - visual inspection, magnification, or quantitative criteria?
Both macro and microscopically
(39) Line 734: We conducted the analysis in three different ways after selecting the spatial window, i.e., the analysed excavated area (52.56 m2).
Clarify why the 52.56 m<sup>2</sup> spatial window was chosen. Was this the total excavated area or a selected portion?
It was what was left of the elephant accumulation after erosion.
(40) Line 728: The spatial statistical analyses of EAK.
Adding one or two sentences at the start explaining the analytical objective, such as testing spatial association between faunal and lithic materials, would help readers understand how each analysis relates to the broader research questions.
This is well explained in the main text
(41) Line 782: An intensive survey seeking stratigraphically-associated megafaunal bones was carried out in the months of June 2023 and 2024.
It would help to specify whether the same areas were resurveyed in both field seasons or if different zones were covered each year. This information is important for understanding sampling consistency and potential spatial bias.
Both areas were surveyed in both field seasons. We were very consistent.
(42) Line 787: We focused on proboscidean bones and used hippopotamus bones, some of the most abundant in the megafaunal fossils, as a spatial control.
Clarify how the hippopotamus remains functional as a "spatial control." Are they used as a proxy for water-associated taxa to test habitat patterning, or as a baseline for comparing carcass distribution? The meaning of "control" in this context is ambiguous.
As a proxy for megafaunal distribution given their greater abundance over any other megafaunal taxa.
(43) Line 789: Stratigraphic association was carried out by direct observation of the geological context and with the presence of a Quaternary geologist during the whole survey.
This is good methodological practice, but it would be helpful to describe how stratigraphic boundaries were identified in the field (for example, by reference to tuffs or marker beds). That information would make the geological framework more replicable.
This is basic geological work. Of course, both tuffs and marker beds were followed.
(44) Line 791: When fossils found were ambiguously associated with specific strata, these were excluded from the present analysis.
You might specify what proportion of the total finds were excluded due to uncertain stratigraphic association. Reporting this would indicate the strength of the stratigraphic control.
This was not quantified but it was a very small amount compared to those whose stratigraphic provenience was certain.
(45) Line 799: The goals of this survey were: a) collect a spatial sample of proboscidean and megafaunal bones enabling us to understand if carcasses on the Olduvai paleolandscapes were randomly deposited or associated to specific habitats.
You might clarify how randomness or habitat association was tested.
Randomness was tested spatially and comparing density according to ecotone. Same for habitat association.
(46) The Methods section provides detailed information about excavation, restoration, and spatial analyses but omits critical details about the zooarchaeological and taphonomic procedures. There is no explanation of how faunal remains were analyzed once recovered, including how cut marks, percussion marks, or green bone fractures were identified or what magnification or diagnostic criteria were used. The authors also do not specify the analytical unit used for faunal quantification (e.g., NISP, MNI, MNE, or other), making it unclear how specimen counts were generated for spatial or taphonomic analyses. Even if these details are provided in the Supplementary Information, the main text should include at least a concise summary describing the analytical framework, the criteria for identifying surface modifications and fracture morphology, and the quantification system employed. This information is essential for transparency, replicability, and proper evaluation of the behavioral interpretations.
See reply above. There is a new subsection on taphonomic methods now.
Supplementary information:
(47) The Supplementary Information includes a large number of green-broken proboscidean specimens from other Olduvai localities (BK, LAS, SC, FLK West), but it is never explained why these are shown or how they relate to the EAK study. The main analysis focuses entirely on the EAK elephant, including so much unrelated material without any stated purpose, which makes the supplement confusing. If these examples are meant only to illustrate the appearance of green fractures, that should be stated. Otherwise, the extensive inclusion of non-EAK material gives the impression that they were part of the analyzed assemblage when they were not.
This is stated in the opening paragraph to the section.
(48) Line 96: A small collection of green-broken elephant bones was retrieved from the lower and upper Bed II units.
It would help to clarify whether these specimens are part of the EAK assemblage or derive from other Bed II localities. As written, it is not clear whether this description refers to material analyzed in the main text or to comparative examples shown only in the Supplementary Information.
No, EAK only occupies the lower Bed II section. They belong in the Bed II paleolandscape units.
(49) Line 97: One of them, a proximal femoral shaft found within the LAS unit, has all the traces of having been used as a tool (Figure 6).
This says the bone tool in Figure 6 is from LAS, but the main text caption identifies it as from EAK. If I am not mistaken, EAK is a site at the base of Bed II, and LAS is a separate stratigraphic unit higher in the sequence, so the authors should clarify which is correct.
Our mistake. It provenience is from LAS in the vicinity of EAK.
(50) Line 186: Figure S20. Example of other megafaunal long bone shafts showing green breaks.
Not cited in text or SI narrative. No indication where these bones come from or why they are relevant.
It appears justified in the revised version.
(51) Line 474: Figure S28-S30. Hyena-ravaged giraffe bones from Chobe (Botswana).
These figures are not discussed in the text or SI, and their relevance to the study is unclear. The authors should explain why these modern comparative examples were included and how they inform interpretations of the Olduvai assemblages.
It appears justified in the revised version.
(52) Line 498: Figure S31. Bos/Bison bone from Bois Roche (France).
This figure is not mentioned in the text or Supplementary Information. The authors should specify why this specimen is shown and how it contributes to the study's taphonomic or behavioral comparisons.
It appears justified in the revised version.
(53) Line 504: Figure S32. Miocene Gomphotherium femur from Spain.
This figure is never referenced in the paper. The authors should clarify the purpose of including a Miocene specimen from outside Africa and explain what it adds to the interpretation of Bed II material.
It appears justified in the revised version.
(54) Line 508: Figure S33. Elephant femoral shaft from BK (Olduvai).
This figure appears to show comparative material but is not cited or discussed in the text. The authors should explain why the BK material is presented here and how it relates to EAK or the broader analysis.
There are two figures labeled S33.
It appears justified in the revised version.
(55) Line 515: Figure S33. Tibia fragment from a large medium-sized bovid displaying multiple overlapping scars on both breakage planes inflicted by carnivore damage.
Because this figure repeats the S33 label and is not cited or explained in the text, it is unclear why this specimen is included or how it contributes to the study. The authors should correct the duplicate numbering and clarify the purpose of this figure.
It appears justified in the revised version.
(56) Line 522: Same specimen as shown in Figure S30, viewed on its medial side.
This is not the same bone as S30. This figure is not discussed in the text or Supplementary Information. The authors should clarify why it is included and how it relates to the rest of the analysis.
It appears justified in the revised version.
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
Public Reviews:
Reviewer #1 (Public review):
Summary:
This study aims to explore how different forms of "fragile nucleosomes" facilitate RNA Polymerase II (Pol II) transcription along gene bodies in human cells. The authors propose that pan-acetylated, pan-phosphorylated, tailless, and combined acetylated/phosphorylated nucleosomes represent distinct fragile states that enable eFicient transcription elongation. Using CUT&Tagseq, RNA-seq, and DRB inhibition assays in HEK293T cells, they report a genome-wide correlation between histone pan-acetylation/phosphorylation and active Pol II occupancy, concluding that these modifications are essential for Pol II elongation.
Strengths:
(1) The manuscript tackles an important and long-standing question about how Pol II overcomes nucleosomal barriers during transcription.
(2) The use of genome-wide CUT&Tag-seq for multiple histone marks (H3K9ac, H4K12ac, H3S10ph, H4S1ph) alongside active Pol II mapping provides a valuable dataset for the community.
(3) The integration of inhibition (DRB) and recovery experiments oFers insight into the coupling between Pol II activity and chromatin modifications.
(4) The concept of "fragile nucleosomes" as a unifying framework is potentially appealing and could stimulate further mechanistic studies.
Really appreciate the positive or affirmative comments from the reviewer.
Weaknesses:
(1) Misrepresentation of prior literature
The introduction incorrectly describes findings from Bintu et al., 2012. The cited work demonstrated that pan-acetylated or tailless nucleosomes reduce the nucleosomal barrier for Pol II passage, rather than showing no improvement. This misstatement undermines the rationale for the current study and should be corrected to accurately reflect prior evidence.
What we said is according to the original report in the publication (Bintu et al., Cell, 2012). Here is the citation from the report:
Page 739,(Bintu, L. et al., Cell, 2012)(PMID: 23141536)
“Overall transcription through tailless and acetylated nucleosomes is slightly faster than through unmodified nucleosomes (Figure 1C), with crossing times that are generally under 1 min (39.5 ± 5.7 and 45.3 ± 7.6 s, respectively). Both the removal and acetylation of the tails increase eFiciency of NPS passage:71% for tailless nucleosomes and 63% for acetylated nucleosomes (Figures 1C and S1), in agreement with results obtained using bulk assays of transcription (Ujva´ ri et al., 2008).”
We will cite this original sentence in our revision.
(2) Incorrect statement regarding hexasome fragility
The authors claim that hexasome nucleosomes "are not fragile," citing older in vitro work. However, recent studies clearly showed that hexasomes exist in cells (e.g., PMID 35597239) and that they markedly reduce the barrier to Pol II (e.g., PMID 40412388). These studies need to be acknowledged and discussed.
“hexasome” was introduced in the transcription field four decades ago. Later, several groups claimed that “hexasome” is fragile and could be generated in transcription elongation of Pol II. However, their original definition was based on the detection of ~100 bps DNA fragments (MNase resistant) in vivo by Micrococcal nuclease sequencing (MNase-seq), which is the right length to wrap up one hexasome histone subunit (two H3/4 and one H2A/2B) to form the sub-nucleosome of a hexasome. As we should all agree that acetylation or phosphorylation of the tails of histone nucleosomes will lead to the compromised interaction between DNA and histone subunits, which could lead to the intact naïve nucleosome being fragile and easy to disassemble, and easy to access by MNase. Fragile nucleosomes lead to better accessibility of MNase to DNA that wraps around the histone octamer, producing shorter DNA fragments (~100 bps instead of ~140 bps). In this regard, we believe that these ~100 bps fragments are the products of fragile nucleosomes (fragile nucleosome --> hexasome), instead of the other way around (hexasome --> fragile).
Actually, two early reports from Dr. David J. Clark’s group from NIH raised questions about the existence of hexasomes in vivo (PMID: 28157509) (PMID: 25348398).
From the report of PMID:35597239, depletion of INO80 leads to the reduction of “hexasome” for a group of genes, and the distribution of both “nucleosomes” and “hexasomes” with the gene bodies gets fuzzier (less signal to noise). In a recent theoretical model (PMID: 41425263), the corresponding PI found that chromatin remodelers could act as drivers of histone modification complexes to carry out different modifications along gene bodies. The PI found that INO80 could drive NuA3 (a H3 acetyltransferase) to carry out pan-acetylation of H3 and possibly H2B as well in the later runs of transcription of Pol II for a group of genes (SAGA-dependent). It suggests that the depletion of INO80 will affect (reduce) the pan-acetylation of nucleosomes, which leads to the drop of pan-acetylated fragile nucleosomes, subsequently the drop of “hexasomes”. This explains why depletion of INO80 leads to the fuzzier results of either nucleosomes or “hexasomes” in PMID: 35597239. The result of PMID: 35597239 could be a strong piece of evidence to support the model proposed by the corresponding PI (PMID: 41425263).
From a recent report: PMID:40412388, the authors claimed that FACT could bind to nucleosomes to generate “hexasomes”, which are fragile for Pol II to overcome the resistance of nucleosomes. It was well established that FACT enhances the processivity of Pol II in vivo via its chaperonin property. However, the exact working mechanism of FACT still remains ambiguous. A report from Dr. Cramer’s group showed that FACT enhances the elongation of regular genes but works just opposite for pausing-regulated genes (PMID: 38810649). An excellent review by Drs. Tim Formosa and Fred Winston showed that FACT is not required for the survival of a group of differentiated cells (PMID: 33104782), suggesting that FACT is not always required for transcription. It is quite tricky to generate naïve hexasomes in vitro according to early reports from the late Dr. Widom’s group. Most importantly, the new data (the speed of Pol II, the best one on bare DNA is ~27 bps/s) from the report of PMID: 40412388, which is much slower than the speed of Pol II in vivo: ~2.5 kbs/min or ~40 bps/s. From our recovering experiments (Fig. 4C, as mentioned by reviewer #3), in 20 minutes (the period between 10 minutes and 30 minutes, due to the property of CUT-&TAG-seq, of which Pol II still active after cells are collected, there is a big delay of complete stop of Pol II during the procedure of CUT&TAG experiments, so the first period of time does not actually reflect the speed of Pol II, which is ~5 kb/min), all Pol IIs move at a uniform speed of ~2.5 kbs/min in vivo. Interestingly, a recent report from Dr. Shixin Liu’s group (PMID: 41310264) showed that adding SPT4/5 to the transcription system with bare DNA (in vitro), the speed of Pol II reaches ~2.5kbs/min, exactly the same one as we derived in vivo. Similar to the original report (PMID: 23141536), the current report of PMID:40412388 does not mimic the conditions in vivo exactly.
There is an urgent need for a revisit of the current definition of “hexasome”, which is claimed to be fragile and could be generated during the elongation of Pol II in vivo. MNase is an enzyme that only works when the substrate is accessible. In inactive regions of the genome, due to the tight packing of chromatin, MNase is not accessible to individual nucleosomes within the bodies of a gene or upstream of promoters, which is why we only see phased/spacing or clear distribution of nucleosomes at the transcription start sites, but it becomes fuzzy downstream or upstream of promoters. On the other hand, for fragile nucleosomes, the accessibility to MNase should increase dramatically, which leads to the ~100 bps fragments. Based on the uniform rate (2.5 kbs/min) of Pol II for all genes derived from human 293T cells and the similar rate (2.5 kbs/min) of Pol II on bare DNA in vitro, it is unlikely for Pol II to pause in the middle of nucleosomes to generate “hexasomes” to continue during elongation along gene bodies. Similar to RNAPs in bacterial (no nucleosomes) and Archaea (tailless nucleosomes), there should be no resistance when Pol IIs transcribe along all fragile nucleosomes within gene bodies in all eukaryotes, as we characterized in this manuscript.
(3) Inaccurate mechanistic interpretation of DRB
The Results section states that DRB causes a "complete shutdown of transcription initiation (Ser5-CTD phosphorylation)." DRB is primarily a CDK9 inhibitor that blocks Pol II release from promoter-proximal pausing. While recent work (PMID: 40315851) suggests that CDK9 can contribute to CTD Ser5/Ser2 di-phosphorylation, the manuscript's claim of initiation shutdown by DRB should be revised to better align with the literature. The data in Figure 4A indicate that 1 M DRB fully inhibits Pol II activity, yet much higher concentrations (10-100 ) are needed to alter H3K9ac and H4K12ac levels. The authors should address this discrepancy by discussing the differential sensitivities of CTD phosphorylation versus histone modification turnover.
Yes, it was reported that DRB is also an inhibitor of CDK9. However, if the reviewer agrees with us and the current view in the field, the phosphorylation of Ser5-CTD of Pol II is the initiation of transcription for all Pol II-regulated genes in eukaryotes. CDK9 is only required to work on the already phosphorylated Ser5-CTD of Pol II to release the paused Pol II, which only happens in metazoans. From a series of works by us and others: CDK9 is unique in metazoans, required only for the pausing-regulated genes but not for regular genes. We found that CDK9 works on initiated Pol II (Ser5-CTD phosphorylated Pol II) and generates a unique phosphorylation pattern on CTD of Pol II (Ser2ph-Ser2ph-Ser5ph-CTD of Pol II), which is required to recruit JMJD5 (via CID domain) to generate a tailless nucleosome at +1 from TSS to release paused Pol II (PMID: 32747552). Interestingly, the report from Dr. Jesper Svejstrup’s group (PMID: 40315851) showed that CDK9 could generate a unique phosphorylation pattern (Ser2ph-Ser5ph-CTD of Pol II), which is not responsive to the popular 3E10 antibody that recognizes the single Ser2phCTD of Pol II. This interesting result is consistent with our early report showing the unique phosphorylation pattern (Ser2ph-Ser2ph-Ser5ph-CTD of Pol II) is specifically generated by CDK9 in animals, which is not recognized by 3E10 either (PMID: 32747552). Actually, an early report from Dr. Dick Eick’s group (PMID: 26799765) showed the difference in the phosphorylation pattern of the CTD of Pol II between animal cells and yeast cells. We have characterized how CDK9 is released from 7SK snRNP and recruited onto paused Pol II via the coupling of JMJD6 and BRD4 (PMID: 32048991), which was published on eLIFE. It is well established that CDK9 works after CDK7 or CDK8. From our PRO-seq data (Fig. 3) and CUT&TAG-seq data of active Pol II (Fig. 4), adding DRB completely shuts down all genes via inhibiting the initiation of Pol II (generation of Ser5ph-CTD of Pol II). Due to the uniqueness of CDK9 only in metazoans, it is not required for the activation of CDK12 or CDK13 (they are orthologs of CTK1 in yeast), as we demonstrated recently (PMID: 41377501). Instead, we found that CDK11/10 acts as the ortholog of Bur1 kinase from yeast, is essential for the phosphorylation of Spt5, the link of CTD of Pol II, and CDK12 (PMID: 41377501).
(4) Insufficient resolution of genome-wide correlations
Figure 1 presents only low-resolution maps, which are Insufficient o determine whether pan-acetylation and pan-phosphorylation correlate with Pol II at promoters or gene bodies. The authors should provide normalized metagene plots (from TSS to TTS) across different subgroups to visualize modification patterns at higher resolution. In addition, the genome-wide distribution of another histone PTM with a diFerent localization pattern should be included as a negative control.
A popular view in the field is that the majority of genomes are inactive since they do not contain coding RNAs, which are responsible for ~20,000 protein candidates characterized in animals. However, our genomewide characterization using the four histone modification marks, active Pol II, and RNA-seq, shows a different story. Figure 1 shows that most of the human genome of HEK293T is active in producing not only protein-coding RNAs but also non-coding RNAs (the majority of them). We believe that Figure 1 could change our current view of the activity of the entire genome, and should be of great interest to general readers as well as researchers on genomics. Furthermore, it is a basis for Figure 2, which is a zoom-in of Figure 1.
(5) Conceptual framing
The manuscript frequently extrapolates correlative genome-wide data to mechanistic conclusions (e.g., that pan-acetylation/phosphorylation "generate" fragile nucleosomes). Without direct biochemical or structural evidence. Such causality statements should be toned down.
The reviewer is right, we should tone down the strong sentences. However, we believe that our data is strong enough to derive the general conclusion. The reviewer may agree with us that the entire field of transcription and epigenetics has been stagnant in recent decades, but there is an urgent need for fresh ideas to change the current situation. Our novel discoveries, for sure, additional supporting data are needed, should open up a brand new avenue for people to explore. We believe that a new era of transcription will emerge based on our novel discoveries. We hope that this manuscript will attract more people to these topics. As Reviewer #3 pointed out, this story establishes the connection between transcription and epigenetics in the field.
Reviewer #2 (Public review):
Summary:
In this manuscript, the authors use various genomics approaches to examine nucleosome acetylation, phosphorylation, and PolII-CTD phosphorylation marks. The results are synthesized into a hypothesis that 'fragile' nucleosomes are associated with active regions of PolII transcription.
Strengths:
The manuscript contains a lot of genome-wide analyses of histone acetylation, histone phosphorylation, and PolII-CTD phosphorylation.
Weaknesses:
This reviewer's main research expertise is in the in vitro study of transcription and its regulation in purified, reconstituted systems.
Actually, the pioneering work of the establishment of in vitro transcription assays at Dr. Robert Roeder’s group led to numerous groundbreaking discoveries in the transcription field. The contributions of in vitro work in the transcription field are the key for us to explore the complexity of transcription in eukaryotes in the early times and remain important currently.
I am not an expert at the genomics approaches and their interpretation, and overall, I had a very hard time understanding and interpreting the data that are presented in this manuscript. I believe this is due to a problem with the manuscript, in that the presentation of the data is not explained in a way that's understandable and interpretable to a non-expert.
Thanks for your suggestions. You are right, we have problems expressing our ideas clearly in this manuscript, which could confuse. We will make modifications accordingly per your suggestions.
For example:
(1) Figure 1 shows genome-wide distributions of H3K9ac, H4K12ac, Ser2phPolII, mRNA, H3S10ph, and H4S1ph, but does not demonstrate correlations/coupling - it is not clear from these data that pan-acetylation and pan-phosphorylation are coupled with Pol II transcription.
Figure 1 shows the overall distribution of the four major histone modifications, active Pol II, and mRNA genome-wide in human HEK293T cells. It tells general readers that the entire genome is quite active and far more than people predicted that most of the genome is inactive, since just a small portion of the genome expresses coding RNAs (~20,000 in animals). Figure 1 shows that the majority of the genome is active and expresses not only coded mRNA but also non-coding RNAs. After all, it is the basis of Figure 2, which is a zoom-in of Figure 1. However, it is beyond the scope of this manuscript to discuss the non-coding RNAs.
(2) Figure 2 - It's not clear to me what Figure 2 is supposed to be showing.
(A) Needs better explanation - what is the meaning of the labels at the top of the gel lanes?
Figure 2 is a zoom-in for the individual gene, which shows how histone modifications are coupled with Pol II activity on the individual gene. We will give a more detailed explanation of the figure per the reviewer’s suggestions.
(B) This reviewer is not familiar with this technique, its visualization, or its interpretation - more explanation is needed. What is the meaning of the quantitation graphs shown at the top? How were these calculated (what is on the y-axis)?
Good suggestions, we will do some modifications.
(3) To my knowledge, the initial observation of DRB eFects on RNA synthesis also concluded that DRB inhibited initiation of RNA chains (pmid:982026) - this needs to be acknowledged.
Thanks for the reference, which is the first report to show the DRB inhibits initiation of Pol II in vivo. We will cite it in the revision.
(4) Again, Figures 4B, 4C, 5, and 6 are very difficult to understand - what is shown in these heat maps, and what is shown in the quantitation graphs on top?
Thanks for the suggestions, we will give a more detailed description of the Figures.
Reviewer #3 (Public review):
Summary:
Li et al. investigated the prevalence of acetylated and phosphorylated histones (using H3K9ac, H4K12ac, H3S10ph & H4S1ph as representative examples) across the gene body of human HEK293T cells, as well as mapping elongating Pol II and mRNA. They found that histone acetylation and phosphorylation were dominant in gene bodies of actively transcribing genes. Genes with acetylation/phosphorylation restricted to the promoter region were also observed. Furthermore, they investigated and reported a correlation between histone modifications and Pol II activity, finding that inhibition of Pol II activity reduced acetylation/phosphorylation levels, while resuming Pol II activity restored them. The authors then proposed a model in which panacetylation or pan-phosphorylation of histones generates fragile nucleosomes; the first round of transcription is accompanied by panacetylation, while subsequent rounds are accompanied by panphosphorylation.
Strengths:
This study addresses a highly significant problem in gene regulation. The author provided riveting evidence that certain histone acetylation and/or phosphorylation within the gene body is correlated with Pol II transcription. The author furthermore made a compelling case that such transcriptionally correlated histone modification is dynamic and can be regulated by Pol II activity. This work has provided a clearer view of the connection between epigenetics and Pol II transcription.
Thanks for the insightful comments, which are exactly what we want to present in this manuscript.
Weaknesses:
The title of the manuscript, "Fragile nucleosomes are essential for RNA Polymerase II to transcribe in eukaryotes", suggests that fragile nucleosomes lead to transcription. While this study shows a correlation between histone modifications in gene bodies and transcription elongation, a causal relationship between the two has not been demonstrated.
Thanks for the suggestions. What we want to express is that the generation of fragile nucleosomes precedes transcription, or, more specifically, transcription elongation. The corresponding PI wrote a hypothetical model on how pan-acetylation is generated by the coupling of chromatin remodelers and acetyltransferase complexes along gene bodies, in which chromatin remodelers act as drivers to carry acetyltransferases along gene bodies to generate pan-acetylation of nucleosomes (PMID: 41425263). We have a series of work to show how “tailless nucleosomes” at +1 from transcription start sites are generated to release paused Pol II in metazoans (PMID: 28847961) (PMID: 29459673) (PMID: 32747552) (PMID: 32048991). We still do not know how pan-phosphorylation along gene bodies is generated. It should be one of the focuses of our future research.
-
-
econweb.ucsd.edu econweb.ucsd.edu
-
Universal pre-school education to support school readiness before first grade
Universal pre-school education aims to ensure that all children enter Grade 1 with basic language, cognitive, and social readiness. In India, this is feasible by strengthening and integrating the existing Anganwadi system with structured early childhood curricula and teacher training. Global examples from Finland, France, and the UK show that universal early childhood education reduces early learning gaps and improves long-term educational outcomes, especially for disadvantaged children.
Why are Anganwadi reforms challenging? 1. Anganwadis were designed for nutrition & care, not education. Hence, the centers are not properly equipped nor is the staff. And if the early schooling is done in an incorrect manner it can lead to a major damage in child's curiosity
- Anganwadi workers are overburdened and undertrained Reality on the ground
An Anganwadi worker today often:
- Handles nutrition distribution
- Maintains health records
- Supports surveys and elections
- Manages multiple government schemes
Now we expect them to: * Teach early literacy * Build number sense * Do classroom management * Track learning progress
Without: * Deep training * Time * Support staff
-
This isn’t resistance - it’s capacity mismatch.
-
Early childhood pedagogy is deceptively hard Biggest misconception that teaching children is easy. This involves knowing how child's brain develops, designing play that secretly builds skills, managing attention spans, language scaffolding through language (Language scaffolding through conversation means helping a child develop language step-by-step by talking with them in a guided way, instead of just teaching words or letters directly.)
-
Coordination problem - Anganwadis operate at the intersection of the women and child development, health, and education systems, but are governed primarily as welfare units rather than educational institutions. As a result, there is no clear ownership or accountability for learning outcomes.
-
-
patrickcollison.com patrickcollison.com
-
but I don’t get the feeling that it’s just that: I think something about authors’ attitudes to the topic changed.
Fitzgerald's stories reference family money situations sometimes. I'm thinking his first novel Tender is the Night but I can't be sure its consistent across the span of his short stories.
Booth Tarkington's The Magnificent Ambersons, while I haven't read the book, initiates the downfall of that family with bad investments by Minaford Sr.
Both aren't detailed about it, sure, but they do continue this trend in a diminished way through works published around the first quarter of the 20th century and I find it interesting their generation was still culturally guided by this motif. As well as all the others of the 19th century novel. Like what another contemporary Edith Wharton accomplished novelistically as well.
-
-
www.biorxiv.org www.biorxiv.org
-
Reviewer #1 (Public review):
Summary:
This study investigates how the brain processes facial expressions across development by analyzing intracranial EEG (iEEG) data from children (ages 5-10) and post-childhood individuals (ages 13-55). The researchers used a short film containing emotional facial expressions and applied AI-based models to decode brain responses to facial emotions. They found that in children, facial emotion information is represented primarily in the posterior superior temporal cortex (pSTC)-a sensory processing area-but not in the dorsolateral prefrontal cortex (DLPFC), which is involved in higher-level social cognition. In contrast, post-childhood individuals showed emotion encoding in both regions. Importantly, the complexity of emotions encoded in the pSTC increased with age, particularly for socially nuanced emotions like embarrassment, guilt, and pride.The authors claim that these findings suggest that emotion recognition matures through increasing involvement of the prefrontal cortex, supporting a developmental trajectory where top-down modulation enhances understanding of complex emotions as children grow older.
Strengths:
(1) The inclusion of pediatric iEEG makes this study uniquely positioned to offer high-resolution temporal and spatial insights into neural development compared to non-invasive approaches, e.g., fMRI, scalp EEG, etc.
(2) Using a naturalistic film paradigm enhances ecological validity compared to static image tasks often used in emotion studies.
(3) The idea of using state-of-the-art AI models to extract facial emotion features allows for high-dimensional and dynamic emotion labeling in real time.
Weaknesses:
(1) The study has notable limitations that constrain the generalizability and depth of its conclusions. The sample size was very small, with only nine children included and just two having sufficient electrode coverage in the posterior superior temporal cortex (pSTC), which weakens the reliability and statistical power of the findings, especially for analyses involving age. Authors pointed out that a similar sample size has been used in previous iEEG studies, but the cited works focus on adults and do not look at the developmental perspectives. Similar work looking at developmental changes in iEEG signals usually includes many more subjects (e.g., n = 101 children from Cross ZR et al., Nature Human Behavior, 2025) to account for inter-subject variabilities.
(2) Electrode coverage was also uneven across brain regions, with not all participants having electrodes in both the dorsolateral prefrontal cortex (DLPFC) and pSTC, making the conclusion regarding the different developmental changes between DLPFC and pSTC hard to interpret (related to point 3 below). It is understood that it is rare to have such iEEG data collected in this age group, and the electrode location is only determined by clinical needs. However, the scientific rigor should not be compromised by the limited data access. It's the authors' decision whether such an approach is valid and appropriate to address the scientific questions, here the developmental changes in the brain, given all the advantages and constraints of the data modality.
(3) The developmental differences observed were based on cross-sectional comparisons rather than longitudinal data, reducing the ability to draw causal conclusions about developmental trajectories. Also, see comments in point 2.
(4) Moreover, the analysis focused narrowly on DLPFC, neglecting other relevant prefrontal areas such as the orbitofrontal cortex (OFC) and anterior cingulate cortex (ACC), which play key roles in emotion and social processing. Agree that this might be beyond the scope of this paper, but a discussion section might be insightful.
(5) Although the use of a naturalistic film stimulus enhances ecological validity, it comes at the cost of experimental control, with no behavioral confirmation of the emotions perceived by participants and uncertain model validity for complex emotional expressions in children. A non-facial music block that could have served as a control was available but not analyzed. The validation of AI model's emotional output needs to be tested. It is understood that we cannot collect these behavioral data retrospectively within the recorded subjects. Maybe potential post-hoc experiments and analyses could be done, e.g., collect behavioral, emotional perception data from age-matched healthy subjects.
(6) Generalizability is further limited by the fact that all participants were neurosurgical patients, potentially with neurological conditions such as epilepsy that may influence brain responses. At least some behavioral measures between the patient population and the healthy groups should be done to ensure the perception of emotions is similar.
(7) Additionally, the high temporal resolution of intracranial EEG was not fully utilized, as data were downsampled and averaged in 500-ms windows. It seems like the authors are trying to compromise the iEEG data analyses to match up with the AI's output resolution, which is 2Hz. It is not clear then why not directly use fMRI, which is non-invasive and seems to meet the needs here already. The advantages of using iEEG in this study are missing here.
(8) Finally, the absence of behavioral measures or eye-tracking data makes it difficult to directly link neural activity to emotional understanding or determine which facial features participants attended to. Related to point 5 as well.
Comments on revisions:
A behavioral measurement will help address a lot of these questions. If the data continues collecting, additional subjects with iEEG recording and also behavioral measurements would be valuable.
-
Author response:
The following is the authors’ response to the original reviews.
eLife Assessment
This study examines a valuable question regarding the developmental trajectory of neural mechanisms supporting facial expression processing. Leveraging a rare intracranial EEG (iEEG) dataset including both children and adults, the authors reported that facial expression recognition mainly engaged the posterior superior temporal cortex (pSTC) among children, while both pSTC and the prefrontal cortex were engaged among adults. However, the sample size is relatively small, with analyses appearing incomplete to fully support the primary claims.
Public Reviews:
Reviewer #1 (Public review):
Summary:
This study investigates how the brain processes facial expressions across development by analyzing intracranial EEG (iEEG) data from children (ages 5-10) and post-childhood individuals (ages 13-55). The researchers used a short film containing emotional facial expressions and applied AI-based models to decode brain responses to facial emotions. They found that in children, facial emotion information is represented primarily in the posterior superior temporal cortex (pSTC) - a sensory processing area - but not in the dorsolateral prefrontal cortex (DLPFC), which is involved in higher-level social cognition. In contrast, post-childhood individuals showed emotion encoding in both regions. Importantly, the complexity of emotions encoded in the pSTC increased with age, particularly for socially nuanced emotions like embarrassment, guilt, and pride. The authors claim that these findings suggest that emotion recognition matures through increasing involvement of the prefrontal cortex, supporting a developmental trajectory where top-down modulation enhances understanding of complex emotions as children grow older.
Strengths:
(1) The inclusion of pediatric iEEG makes this study uniquely positioned to offer high-resolution temporal and spatial insights into neural development compared to non-invasive approaches, e.g., fMRI, scalp EEG, etc.
(2) Using a naturalistic film paradigm enhances ecological validity compared to static image tasks often used in emotion studies.
(3) The idea of using state-of-the-art AI models to extract facial emotion features allows for high-dimensional and dynamic emotion labeling in real time
Weaknesses:
(1) The study has notable limitations that constrain the generalizability and depth of its conclusions. The sample size was very small, with only nine children included and just two having sufficient electrode coverage in the posterior superior temporal cortex (pSTC), which weakens the reliability and statistical power of the findings, especially for analyses involving age
We appreciated the reviewer’s point regarding the constrained sample size.
As an invasive method, iEEG recordings can only be obtained from patients undergoing electrode implantation for clinical purposes. Thus, iEEG data from young children are extremely rare, and rapidly increasing the sample size within a few years is not feasible. However, we are confident in the reliability of our main conclusions. Specifically, 8 children (53 recording contacts in total) and 13 control participants (99 recording contacts in total) with electrode coverage in the DLPFC are included in our DLPFC analysis. This sample size is comparable to other iEEG studies with similar experiment designs [1-3].
For pSTC, we returned to the data set and found another two children who had pSTC coverage. After involving these children’s data, the group-level analysis using permutation test showed that children’s pSTC significantly encode facial emotion in naturalistic contexts (Figure 3B). Notably, the two new children’s (S33 and S49) responses were highly consistent with our previous observations. Moreover, the averaged prediction accuracy in children’s pSTC (r<sub>speech</sub>=0.1565) was highly comparable to that in post-childhood group (r<sub>speech</sub>=0.1515).
(1) Zheng, J. et al. Multiplexing of Theta and Alpha Rhythms in the Amygdala-Hippocampal Circuit Supports Pafern Separation of Emotional Information. Neuron 102, 887-898.e5 (2019).
(2) Diamond, J. M. et al. Focal seizures induce spatiotemporally organized spiking activity in the human cortex. Nat. Commun. 15, 7075 (2024).
(3) Schrouff, J. et al. Fast temporal dynamics and causal relevance of face processing in the human temporal cortex. Nat. Commun. 11, 656 (2020).
(2) Electrode coverage was also uneven across brain regions, with not all participants having electrodes in both the dorsolateral prefrontal cortex (DLPFC) and pSTC, and most coverage limited to the left hemisphere-hindering within-subject comparisons and limiting insights into lateralization.
The electrode coverage in each patient is determined entirely by the clinical needs. Only a few patients have electrodes in both DLPFC and pSTC because these two regions are far apart, so it’s rare for a single patient’s suspected seizure network to span such a large territory. However, it does not affect our results, as most iEEG studies combine data from multiple patients to achieve sufficient electrode coverage in each target brain area. As our data are mainly from left hemisphere (due to the clinical needs), this study was not designed to examine whether there is a difference between hemispheres in emotion encoding. Nevertheless, lateralization remains an interesting question that should be addressed in future research, and we have noted this limitation in the Discussion (Page 8, in the last paragraph of the Discussion).
(3) The developmental differences observed were based on cross-sectional comparisons rather than longitudinal data, reducing the ability to draw causal conclusions about developmental trajectories.
In the context of pediatric intracranial EEG, longitudinal data collection is not feasible due to the invasive nature of electrode implantation. We have added this point to the Discussion to acknowledge that while our results reveal robust age-related differences in the cortical encoding of facial emotions, longitudinal studies using non-invasive methods will be essential to directly track developmental trajectories (Page 8, in the last paragraph of Discussion). In addition, we revised our manuscript to avoid emphasis causal conclusions about developmental trajectories in the current study (For example, we use “imply” instead of “suggest” in the fifth paragraph of Discussion).
(4) Moreover, the analysis focused narrowly on DLPFC, neglecting other relevant prefrontal areas such as the orbitofrontal cortex (OFC) and anterior cingulate cortex (ACC), which play key roles in emotion and social processing.
We agree that both OFC and ACC are critically involved in emotion and social processing. However, we have no recordings from these areas because ECoG rarely covers the ACC or OFC due to technical constraints. We have noted this limitation in the Discussion(Page 8, in the last paragraph of Discussion). Future follow-up studies using sEEG or non-invasive imaging methods could be used to examine developmental patterns in these regions.
(5) Although the use of a naturalistic film stimulus enhances ecological validity, it comes at the cost of experimental control, with no behavioral confirmation of the emotions perceived by participants and uncertain model validity for complex emotional expressions in children. A nonfacial music block that could have served as a control was available but not analyzed.
The facial emotion features used in our encoding models were extracted by Hume AI models, which were trained on human intensity ratings of large-scale, experimentally controlled emotional expression data[1-2]. Thus, the outputs of Hume AI model reflect what typical facial expressions convey, that is, the presented facial emotion. Our goal of the present study was to examine how facial emotions presented in the videos are encoded in the human brain at different developmental stages. We agree that children’s interpretation of complex emotions may differ from that of adults, resulting in different perceived emotion (i.e., the emotion that the observer subjectively interprets). Behavioral ratings are necessary to study the encoding of subjectively perceived emotion, which is a very interesting direction but beyond the scope of the present work. We have added a paragraph in the Discussion (see Page 8) to explicitly note that our study focused on the encoding of presented emotion.
We appreciated the reviewer’s point regarding the value of non-facial music blocks. However, although there are segments in music condition that have no faces presented, these cannot be used as a control condition to test whether the encoding model’s prediction accuracy in pSTC or DLPFC drops to chance when no facial emotion is present. This is because, in the absence of faces, no extracted emotion features are available to be used for the construction of encoding model (see Author response image 1 below). Thus, we chose to use a different control analysis for the present work. For children’s pSTC, we shuffled facial emotion feature in time to generate a null distribution, which was then used to test the statistical significance of the encoding models (see Methods/Encoding model fitting for details).
(1) Brooks, J. A. et al. Deep learning reveals what facial expressions mean to people in different cultures. iScience 27, 109175 (2024).
(2) Brooks, J. A. et al. Deep learning reveals what vocal bursts express in different cultures. Nat. Hum. Behav. 7, 240–250 (2023).
Author response image 1.
Time courses of Hume AI extracted facial expression features for the first block of music condition. Only top 5 facial expressions were shown here to due to space limitation.
(6) Generalizability is further limited by the fact that all participants were neurosurgical patients, potentially with neurological conditions such as epilepsy that may influence brain responses.
We appreciated the reviewer’s point. However, iEEG data can only be obtained from clinical populations (usually epilepsy patients) who have electrodes implantation. Given current knowledge about focal epilepsy and its potential effects on brain activity, researchers believe that epilepsy-affected brains can serve as a reasonable proxy for normal human brains when confounding influences are minimized through rigorous procedures[1]. In our study, we took several steps to ensure data quality: (1) all data segments containing epileptiform discharges were identified and removed at the very beginning of preprocessing, (2) patients were asked to participate the experiment several hours outside the window of seizures. Please see Method for data quality check description (Page 9/ Experimental procedures and iEEG data processing).
(1) Parvizi J, Kastner S. 2018. Promises and limitations of human intracranial electroencephalography. Nat Neurosci 21:474–483. doi:10.1038/s41593-018-0108-2
(7) Additionally, the high temporal resolution of intracranial EEG was not fully utilized, as data were down-sampled and averaged in 500-ms windows.
We agree that one of the major advantages of iEEG is its millisecond-level temporal resolution. In our case, the main reason for down-sampling was that the time series of facial emotion features extracted from the videos had a temporal resolution of 2 Hz, which were used for the modelling neural responses. In naturalistic contexts, facial emotion features do not change on a millisecond timescale, so a 500 ms window is sufficient to capture the relevant dynamics. Another advantage of iEEG is its tolerance to motion, which is excessive in young children (e.g., 5-year-olds). This makes our dataset uniquely valuable, suggesting robust representation in the pSTC but not in the DLPFC in young children. Moreover, since our method framework (Figure 1) does not rely on high temporal resolution method, so it can be transferred to non-invasive modalities such as fMRI, enabling future studies to test these developmental patterns in larger populations.
(8) Finally, the absence of behavioral measures or eye-tracking data makes it difficult to directly link neural activity to emotional understanding or determine which facial features participants afended to.
We appreciated this point. Part of our rationale is presented in our response to (5) for the absence of behavioral measures. Following the same rationale, identifying which facial features participants attended to is not necessary for testing our main hypotheses because our analyses examined responses to the overall emotional content of the faces. However, we agree and recommend future studies use eye-tracking and corresponding behavioral measures in studies of subjective emotional understanding.
Reviewer #2 (Public review):
Summary:
In this paper, Fan et al. aim to characterize how neural representations of facial emotions evolve from childhood to adulthood. Using intracranial EEG recordings from participants aged 5 to 55, the authors assess the encoding of emotional content in high-level cortical regions. They report that while both the posterior superior temporal cortex (pSTC) and dorsolateral prefrontal cortex (DLPFC) are involved in representing facial emotions in older individuals, only the pSTC shows significant encoding in children. Moreover, the encoding of complex emotions in the pSTC appears to strengthen with age. These findings lead the authors to suggest that young children rely more on low-level sensory areas and propose a developmental shiZ from reliance on lower-level sensory areas in early childhood to increased top-down modulation by the prefrontal cortex as individuals mature.
Strengths:
(1) Rare and valuable dataset: The use of intracranial EEG recordings in a developmental sample is highly unusual and provides a unique opportunity to investigate neural dynamics with both high spatial and temporal resolution.
(2) Developmentally relevant design: The broad age range and cross-sectional design are well-suited to explore age-related changes in neural representations.
(3) Ecological validity: The use of naturalistic stimuli (movie clips) increases the ecological relevance of the findings.
(4) Feature-based analysis: The authors employ AIbased tools to extract emotion-related features from naturalistic stimuli, which enables a data-driven approach to decoding neural representations of emotional content. This method allows for a more fine-grained analysis of emotion processing beyond traditional categorical labels.
Weaknesses:
(1) The emotional stimuli included facial expressions embedded in speech or music, making it difficult to isolate neural responses to facial emotion per se from those related to speech content or music-induced emotion.
We thank the reviewer for their raising this important point. We agree that in naturalistic settings, face often co-occur with speech, and that these sources of emotion can overlap. However, background music induced emotions have distinct temporal dynamics which are separable from facial emotion (See the Author response image 2 (A) and (B) below). In addition, face can convey a wide range of emotions (48 categories in Hume AI model), whereas music conveys far fewer (13 categories reported by a recent study [1]). Thus, when using facial emotion feature time series as regressors (with 48 emotion categories and rapid temporal dynamics), the model performance will reflect neural encoding of facial emotion in the music condition, rather than the slower and lower-dimensional emotion from music.
For the speech condition, we acknowledge that it is difficult to fully isolate neural responses to facial emotion from those to speech when the emotional content from faces and speech highly overlaps. However, in our study, (1) the time courses of emotion features from face and voice are still different (Author response image 2 (C) and (D)), (2) our main finding that DLPFC encodes facial expression information in postchildhood individuals but not in young children was found in both speech and music condition (Figure 2B and 2C). In music condition, neural responses to facial emotion are not affected by speech. Thus, we have included the DLPFC results from the music condition in the revised manuscript (Figure 2C), and we acknowledge that this issue should be carefully considered in future studies using videos with speech, as we have indicated in the future directions in the last paragraph of Discussion.
(1) Cowen, A. S., Fang, X., Sauter, D. & Keltner, D. What music makes us feel: At least 13 dimensions organize subjective experiences associated with music across different cultures. Proc Natl Acad Sci USA 117, 1924–1934 (2020).
Author response image 2.
Time courses of the amusement. (A) and (B) Amusement conveyed by face or music in a 30-s music block. Facial emotion features are extracted by Hume AI. For emotion from music, we approximated the amusement time course using a weighted combination of low-level acoustic features (RMS energy, spectral centroid, MFCCs), which capture intensity, brightness, and timbre cues linked to amusement. Notice that music continues when there are no faces presented. (C) and (D) Amusement conveyed by face or voice in a 30-s speech block. From 0 to 5 seconds, a girl is introducing her friend to a stranger. The camera focuses on the friend, who appears nervous, while the girl’s voice sounds cheerful. This mismatch explains why the shapes of the two time series differ at the beginning. Such situations occur frequently in naturalistic movies
(2) While the authors leveraged Hume AI to extract facial expression features from the video stimuli, they did not provide any validation of the tool's accuracy or reliability in the context of their dataset. It remains unclear how well the AI-derived emotion ratings align with human perception, particularly given the complexity and variability of naturalistic stimuli. Without such validation, it is difficult to assess the interpretability and robustness of the decoding results based on these features.
Hume AI models were trained and validated by human intensity ratings of large-scale, experimentally controlled emotional expression data [1-2]. The training process used both manual annotations from human raters and deep neural networks. Over 3000 human raters categorized facial expressions into emotion categories and rated on a 1-100 intensity scale. Thus, the outputs of Hume AI model reflect what typical facial expressions convey (based on how people actually interpret them), that is, the presented facial emotion. Our goal of the present study was to examine how facial emotions presented in the videos are encoded in the human brain at different developmental stages. We agree that the interpretation of facial emotions may be different in individual participants, resulting in different perceived emotion (i.e., the emotion that the observer subjectively interprets). Behavioral ratings are necessary to study the encoding of subjectively perceived emotion, which is a very interesting direction but beyond the scope of the present work. We have added text in the Discussion to explicitly note that our study focused on the encoding of presented emotion (second paragraph in Page 8).
(1) Brooks, J. A. et al. Deep learning reveals what facial expressions mean to people in different cultures. iScience 27, 109175 (2024).
(2) Brooks, J. A. et al. Deep learning reveals what vocal bursts express in different cultures. Nat. Hum. Behav. 7, 240–250 (2023).
(3) Only two children had relevant pSTC coverage, severely limiting the reliability and generalizability of results.
We appreciated this point and agreed with both reviewers who raised it as a significant concern. As described in response to reviewer 1 (comment 1), we have added data from another two children who have pSTC coverage. Group-level analysis using permutation test showed that children’s pSTC significantly encode facial emotion in naturalistic contexts (Figure 3B). Because iEEG data from young children are extremely rare, rapidly increasing the sample size within a few years is not feasible. However, we are confident in the reliability of our conclusion that children’s pSTC can encode facial emotion. First, the two new children’s responses (S33 and S49) from pSTC were highly consistent with our previous observations (see individual data in Figure 3B). Second, the averaged prediction accuracy in children’s pSTC (r<sub>speech</sub>=0.1565) was highly comparable to that in post-childhood group (r<sub>speech</sub>=0.1515).
(4) The rationale for focusing exclusively on high-frequency activity for decoding emotion representations is not provided, nor are results from other frequency bands explored.
We focused on high-frequency broadband (HFB) activity because it is widely considered to reflect the responses of local neuronal populations near the recording electrode, whereas low-frequency oscillations in the theta, alpha, and beta ranges are thought to serve as carrier frequencies for long-range communication across distributed networks[1-2]. Since our study aimed to examine the representation of facial emotion in localized cortical regions (DLPFC and pSTC), HFB activity provides the most direct measure of the relevant neural responses. We have added this rationale to the manuscript (Page 3).
(1) Parvizi, J. & Kastner, S. Promises and limitations of human intracranial electroencephalography. Nat. Neurosci. 21, 474–483 (2018).
(2) Buzsaki, G. Rhythms of the Brain. (Oxford University Press, Oxford, 200ti).
(5) The hypothesis of developmental emergence of top-down prefrontal modulation is not directly tested. No connectivity or co-activation analyses are reported, and the number of participants with simultaneous coverage of pSTC and DLPFC is not specified.
Directional connectivity analysis results were not shown because only one child has simultaneous coverage of pSTC and DLPFC. However, the Granger Causality results from post-childhood group (N=7) clearly showed that the influence in the alpha/beta band from DLPFC to pSTC (top-down) is gradually increased above the onset of face presentation (Author response image 3, below left, plotted in red). By comparison, the influence in the alpha/beta band from pSTC to DLPFC (bottom-up) is gradually decreased after the onset of face presentation (Author response image 3, below left, blue curve). The influence in alpha/beta band from DLPFC to pSTC was significantly increased at 750 and 1250 ms after the face presentation (face vs nonface, paired t-test, Bonferroni corrected P=0.005, 0.006), suggesting an enhanced top-down modulation in the post-childhood group during watching emotional faces. Interestingly, this top-down influence appears very different in the 8-year-old child at 1250 ms after the face presentation (Author response image 3, below left, black curve).
As we cannot draw direct conclusions from the single-subject sample presented here, the top-down hypothesis is introduced only as a possible explanation for our current results. We have removed potentially misleading statements, and we plan to test this hypothesis directly using MEG in the future.
Author response image 3.
Difference of Granger causality indices (face – nonface) in alpha/beta and gamma band for both directions. We identified a series of face onset in the movie that paticipant watched. Each trial was defined as -0.1 to 1.5 s relative to the onset. For the non-face control trials, we used houses, animals and scenes. Granger causality was calculated for 0-0.5 s, 0.5-1 s and 1-1.5 s time window. For the post-childhood group, GC indices were averaged across participants. Error bar is sem.
(6) The "post-childhood" group spans ages 13-55, conflating adolescence, young adulthood, and middle age. Developmental conclusions would benefit from finer age stratification.
We appreciate this insightful comment. Our current sample size does not allow such stratification. But we plan to address this important issue in future MEG studies with larger cohorts.
(7) The so-called "complex emotions" (e.g., embarrassment, pride, guilt, interest) used in the study often require contextual information, such as speech or narrative cues, for accurate interpretation, and are not typically discernible from facial expressions alone. As such, the observed age-related increase in neural encoding of these emotions may reflect not solely the maturation of facial emotion perception, but rather the development of integrative processing that combines facial, linguistic, and contextual cues. This raises the possibility that the reported effects are driven in part by language comprehension or broader social-cognitive integration, rather than by changes in facial expression processing per se.
We agree with this interpretation. Indeed, our results already show that speech influences the encoding of facial emotion in the DLPFC differently in the childhood and post-childhood groups (Figure 2D), suggesting that children’s ability to integrate multiple cues is still developing. Future studies are needed to systematically examine how linguistic cues and prior experiences contribute to the understanding of complex emotions from faces, which we have added to our future directions section (last paragraph in Discussion, Page 8-9 ).
Recommendations for the authors:
Reviewer #1 (Recommendations for the authors):
In the introduction: "These neuroimaging data imply that social and emotional experiences shape the prefrontal cortex's involvement in processing the emotional meaning of faces throughout development, probably through top-down modulation of early sensory areas." Aren't these supposed to be iEEG data instead of neuroimaging?
Corrected.
Reviewer #2 (Recommendations for the authors):
This manuscript would benefit from several improvements to strengthen the validity and interpretability of the findings:
(1) Increase the sample size, especially for children with pSTC coverage.
We added data from another two children who have pSTC coverage. Please see our response to reviewer 2’s comment 3 and reviewer 1’s comment 1.
(2) Include directional connectivity analyses to test the proposed top-down modulation from DLPFC to pSTC.
Thanks for the suggestion. Please see our response to reviewer 2’s comment 5.
(3) Use controlled stimuli in an additional experiment to separate the effects of facial expression, speech, and music.
This is an excellent point. However, iEEG data collection from children is an exceptionally rare opportunity and typically requires many years, so we are unable to add a controlled-stimulus experiment to the current study. We plan to consider using controlled stimuli to study the processing of complex emotion using non-invasive method in the future. In addition, please see our response to reviewer 2’s comment 1 for a description of how neural responses to facial expression and music are separated in our study.
-
-
Local file Local file
-
Balogh’s adoption of not just onebut many racial identities demonstrates the flexibility with which whitepeople take on other identities, often in the name of valuing or “respect-ing” them. This availability, second, underwrites the fluidity of relations ofidentification between white individuals and race, in that, in contrast tothose who are racially interpellated, the basis of these relations is a matterof individual determination; the basis of racial identity can be less a matterof imposed categories than subjective selection. Thereby, third, thoserelations themselves are disposable or, alternately, inalienable.
Owing to the body-mind duality myth, that is. This logical argument chain makes sense insofar as people could perform as black and then be black, much in line with TERF anti-trans ideologies. Yet, this gets rid of the complexities of the process and accumulated lived experiences, basically strawmaning "I want to be a girl" or "I must abort" as if they were trends.
Now, even if instrumentally acknowledging it as an aesthetic-identity privilege "entitlement" of some white people who transition (though note most are vulnerable and face many barriers, institutional, economical, and familiar, from an already poor household), the logic disregards the impacts of trends and long term, only focusing on the immediate after. It fearmongs in an infantilising about the "correct (hegemonic) development" for an adolescent, but most importantly, it decenters the loss of privilege and the abuse and backlash faced from following this process, highligting the statist objectivist biases that engendered binaryist stereotypes.
One can't simply impersonate and live someone else's life. Fantasy can approach us to it, and be very vivid at doing so, but the approach will be a momentary peak of attention that is not usually maintained as it is not continuously lived, embodied.
Identity might not be a right, sure, but oftentimes, but the right-wind reframing displaces the debate here... we are not visiting Jakarta or the North Pole as exotic tourists, we are commiting to a change of reality because over multiple years of questioning and trying changes, we've decided it's the way to go. This is not about white people having a right to racialised bodies like fast clothing or food, this is about acknoweldging other ways of living as valid through an ethics of care that decenters the falsely "efficient" white male default. It's about saying yes to diverse ways of living that don't exploit the world, because some "being" blind, asexual, autistic, indigenous, and why not, fat, is not somethign that one usually "choses", and thereby, without relying on naturalistic fallacies but on a decentering anti-monopoly axiology, we should provide safe spaces for these to flourish.
-
Logged in as “Dead_in_Iraq,” DeLappe types the names of soldiers killed in Iraq, andthe date of their death, into the game’s text messaging system,such that the information scrolls across the screen for all users tosee. DeLappe’s goal is simple: He plans to memorialize the nameof every service member killed in Iraq.
I hope it's not just American soldiers... and wydm just soldiers? If This War of Mine showed us something, it's that soldiers are not the only victims of war.
-
-
www.biorxiv.org www.biorxiv.org
-
Reviewer #1 (Public review):
Summary
The strength of this manuscript lies in the behavior: mice use a continuous auditory background (pink vs brown noise) to set a rule for interpreting an identical single-whisker deflection (lick in W+ and withhold in W− contexts) while always licking to a brief 10 kHz tone. Behaviorally, animals acquire the rule and switch rapidly at block transitions and take a few trials to fully integrate the context cue. What's nice about this behavior is the separate auditory cue, which shows the animals remain engaged in the task, so it's not just that the mice check out (i.e., become disengaged in the W- context). The authors then use optical tools, combining cortex-wide optogenetic inactivation (using localized inhibition in a grid-like fashion) with widefield calcium imaging to map what regions are necessary for the task and what the local and global dynamics are. Classic whisker sensorimotor nodes (wS1/wS2/wM/ALM) behave as expected with silencing reducing whisker-evoked licking. Retrosplenial cortex (RSC) emerges as a somewhat unexpected, context-specific node: silencing RSC (and tjS1) increases licking selectively in W−, arguing that these regions contribute to applying the "don't lick" policy in that context. I say somewhat because work from the Delamater group points to this possibility, albeit in a Pavlovian conditioning task and without neural data. I would still recommend the authors of the current manuscript review that work to see whether there is a relevant framework or concept (Castiello, Zhang, Delamater, 'The retrosplenial cortex as a possible 'sensory integration' area: a neural network modeling approach of the differential outcomes effect of negative patterning', 2021, Neurobiology of Learning and Memory).
The widefield imaging shows that RSC is the earliest dorsal cortical area to show W+ vs W− divergence after the whisker stimulus, preceding whisker motor cortex, consistent with RSC injecting context into the sensorimotor flow. A "Context Off" control (continuous white noise; same block structure) impairs context discrimination, indicating the continuous background is actually used to set the rule (an important addition!) Pre-stimulus functional-connectivity analyses suggest that there is some activity correlation that maps to the context presumably due to the continuous background auditory context. Simultaneous opto+imaging projects perturbations into a low-dimensional subspace that separates lick vs no-lick trajectories in an interpretable way.
In my view, this is a clear, rigorous systems-level study that identifies an important role for RSC in context-dependent sensorimotor transformation, thereby expanding RSC's involvement beyond navigation/memory into active sensing and action selection. The behavioral paradigm is thoughtfully designed, the claims related to the imaging are well defended, and the causal mapping is strong. I have a few suggestions for clarity that may require a bit of data analysis. I also outline one key limitation that should be discussed, but is likely beyond the scope of this manuscript.
Major strengths
(1) The task is a major strength. It asks the animal to generate differential motor output to the same sensory stimulus, does so in a block-based manner, and the Context-Off condition convincingly shows that the continuous contextual cue is necessary. The auditory tone control ensures this is more than a 'motivational' context but is decision-related. In fact, the slightly higher bias to lick on the catch trials in the W+ context is further evidence for this.
(2) The dorsal-cortex optogenetic grid avoids a 'look-where-we-expect' approach and lets RSC fall out as a key node. The authors then follow this up with pharmacology and latency analyses to rule out simple motor confounds. Overall, this is rigorous and thoughtfully done.
(3) While the mesoscale imaging doesn't allow for cellular resolution, it allows for mapping of the flow of information. It places RSC early in the context-specific divergence after whisker onset, a valuable piece that complements prior work.
(4) The baseline (pre-stim) functional connectivity and the opto-perturbation projections into a task subspace increase the significance of the work by moving beyond local correlates.
Key limitation
The current optogenetic window begins ~10 ms before the sensory cue and extends 1s after, which is ideal for perturbing within-trial dynamics but cannot isolate whether RSC is required to maintain the context-specific rule during the baseline. Because context is continuously available, it makes me wonder whether RSC is the locus maintaining or, instead, gating the context signal. The paper's results are fully consistent with that possibility, but causality in the pre-stimulus window remains an open question. (As a pointer for future work, pre-stimulus-only inactivation, silencing around block switches, or context-omission probe trials (e.g., removing the background noise unexpectedly within a W+ or W- context block), could help separate 'holding' from 'gating' of the rule. But I'm not suggesting these are needed for this manuscript, but would be interesting for future studies.)
-
-
-
Free to Download Sweet Venom (Vipers, #2)
by Rina Kent
Download Now
Overview :
From the New York Times & USA Today bestselling author Rina Kent comes a dangerously dark stalker hockey romance.Can I outrun his merciless obsession?I accidentally witnessed a brutal murder.I froze, pretended I saw nothing, hoping I could leave it behind.But my plan backfired, and my life spiraled downward.Now, I’m the target of cold-blooded revenge.Jude Callahan isn’t just a hockey god—he’s a devil no one dares to cross.My existence disrupts his stardom, prestige, and possible serial killer career choice.And he’s set out to make me pay for that moment of silence.No matter how much I run or hide, he finds me, watching from the shadows.Like a predator.I thought he’d stop at the stalking.Or even better, he’d kill me and finally end my misery.But Jude has other plans.He says I can’t die. I have to pay for my sins.And just like that, he drags me into his depraved world, kicking and screaming.This book can be read on its own but for better understanding of the world, it's recommended to read Beautiful Venom first. The pacing of the book reflects a careful consideration of reader engagement. Moments of intensity are balanced with quieter scenes that provide context and reflection. This rhythm keeps the story dynamic without becoming overwhelming. This book stands out for its ability to convey complex ideas in a way that feels natural and easy to follow. Rather than relying on excessive exposition, the author allows the story to evolve organically through dialogue and action. Readers often appreciate how the pacing remains consistent, avoiding unnecessary detours while still providing enough detail to fully understand the motivations behind each character. This approach creates a reading experience that feels both thoughtful and satisfying. The narrative voice used throughout the book feels confident and well-defined. The author’s tone remains steady, helping readers develop a sense of familiarity with the storytelling style. This consistency makes it easier to follow the plot and understand the underlying messages woven into the text. The result is a cohesive reading experience that feels deliberate and carefully crafted.
Include Format in : √pdf, √e-book, √book, √ePub
-
-
www.scratchapixel.com www.scratchapixel.comGeometry2
-
If B is a unit vector, the operation A⋅B yields ||A||cos(θ), signifying the magnitude of A's projection in B's direction, with a negative sign if the direction is reversed. This is termed the scalar projection of A onto B.
Chat with Gemini:<br /> Lee:
I'm taking an online course of 3D graphics. Geometry (as it pertains to 3D graphics on computers) is the current focus. The text just said:
If B is a unit vector, the operation A⋅B yields ∥A∥cos(θ), signifying the magnitude of A's projection in B's direction, with a negative sign if the direction is reversed. This is termed the scalar projection of A onto B. Can you explain this better to me? What does "magnitude of A's (a non-unit vector) projection in B's (a unit vector) direction " mean?
Gemini:<br /> This is one of the most fundamental concepts in 3D graphics, but the math jargon can sometimes obscure the simple visual reality.
To understand "the magnitude of A's projection in B's direction," the best tool is the Shadow Analogy.
1. The Shadow Analogy
Imagine Vector A is a stick floating in space. Vector B represents the ground (or a specific direction along the ground).
If you were to shine a light directly from above (perpendicular to the ground B), the stick (A) would cast a shadow onto the ground.
- The Projection: The "shadow" itself is the projection.
- The Magnitude: The length of that shadow is the "magnitude of the projection."
So, when the text says "magnitude of A's projection in B's direction," it is simply asking: "How much of Vector A is pointing in the same direction as Vector B?"
2. Why does B have to be a Unit Vector?
This is the crucial part that often trips people up.
In 3D graphics, a Unit Vector has a length of exactly 1. It is used purely to point the way; it contains direction information but no "scale" or "weight."
The formula for the dot product is:
** MISSING FORMULAS HERE **
If B is a unit vector, then . The formula simplifies to:
If B wasn't a unit vector (say, it was 5 units long), the result would be the length of the shadow multiplied by 5. That gives you a useless number if you just wanted to measure the shadow. By ensuring B is length 1, the math gives you the exact, unscaled length of the shadow of A.
3. Visualizing the Angles (The "Negative" Sign)
The text mentioned a "negative sign if the direction is reversed." This happens based on the angle () between the two vectors.
- Acute Angle (): The vectors point generally in the same direction. The shadow falls forward along B. The result is Positive.
- Right Angle (): The vectors are perpendicular. There is no shadow (it's just a dot). The result is Zero.
- Obtuse Angle (): The vectors point generally away from each other. The shadow falls backward behind the start of B. The result is Negative.
Summary Table
``` | Concept | The Visual | The Math | | ----------------- | ---------------------------------------- | ------------- | | Vector A | The object casting the shadow. | ∥A∥ | | Vector B (Unit) | The ground/direction we measure against. | ∥B∥=1 | | Dot Product (A⋅B) | The length of the shadow. | A⋅B=∥A∥cos(θ) |
```
Why do you need this in 3D Graphics?
You will use this constantly. Here are two immediate examples:
- Lighting: If B is the surface normal (pointing out of a polygon) and A is the light direction, the dot product tells you how directly the light hits the surface. High projection = Bright light. Negative projection = The light is hitting the back of the polygon (it's dark).<br />
- Mario running up a slope: If B is the direction of the slope and A is Mario's velocity, the projection tells you how fast he is actually moving up the hill vs just moving forward.
Lee:
So if B was pointing directly along the +X axis (X=1, Y=0, Z=0) and A was angled up from that and longer (X=2, Y=2, Z=0), A's magnitude would be ~2.83. But A⋅B would be 2 (It's X coordinate in this specific scenario). If you made a triangle with A being one side and B specifying the direction of the adjacent side, and drew a straight line from the end of A straight down to and perpendicular to the direction B, the length of that "B" side would be A⋅B assuming ∥B∥ was 1. Does that sound correct?
Gemini:<br /> Yes, that is exactly correct. You have perfectly grasped the geometric interpretation of the dot product.
Your example scenario is spot on. Here is the breakdown of why your visualization works, mapping your description to the standard trigonometry terms.
1. The Math Check
Using your numbers:
- Vector A: (Magnitude )
- Vector B: (Unit vector along X)
The calculation is:
You correctly identified that because B is aligned perfectly with the X-axis, the "shadow" of A is simply its X-coordinate (2).
2. The Triangle Visualization
You described making a triangle. This is the exact geometric definition used in physics and graphics.
- The Hypotenuse: This is your Vector A (length ).
- The "Drop" Line: The line you drew "straight down to and perpendicular to B" is the opposite side of the triangle.
- The Adjacent Side: The side lying along the direction of B is the Scalar Projection. Its length is exactly the dot product (2).
In trigonometry terms:
3. One Crucial Detail
You added the condition: "assuming ∥B∥ was 1."
This is the most important part of your insight. If Vector B had been (2, 0, 0) (length of 2), the dot product would have been:
(2×2) + (2×0) + (0×0) = 4The math would return 4, but the "shadow" is physically still only 2 units long. The dot product scales the result by the magnitude of B. That is why we must normalize B (make it a unit vector) before using it as a "measuring stick."
You have the concept of Scalar Projection (the length of the shadow) down cold.
-
When both vectors are normalized, the arc cosine (cos−1) of their dot product reveals the angle θ between them: θ=cos−1(A⋅B||A||||B||) or θ=cos−1(A^⋅B^), where cos−1 denotes the inverse cosine function, commonly represented as acos() in programming languages.
Chat with Gemini:<br /> Lee:
Here's another block of text in that same lesson. Can you help me visualize what it means?
When both vectors are normalized, the arc cosine (cos⁻¹) of their dot product reveals the angle θ between them: θ=cos⁻¹((A⋅B)/(∥A∥ ∥B∥)) or cos⁻¹(Â⋅B̂), where cos⁻¹ denotes the inverse cosine function, commonly represented as acos() in programming languages.
Gemini:<br /> This block of text is explaining how to "reverse-engineer" an angle using the dot product. In 3D graphics, you often know the coordinates of two objects (like a player and an enemy), but you don't know the angle between them. This formula is how you find it.
The Logic: Working Backwards
Think of the dot product formula we just discussed:
A⋅B = ∥A∥∥B∥cos(θ)The text is simply rearranging that equation to solve for .
- Isolate the Cosine: Divide both sides by the magnitudes ().
A⋅B cos(θ) = ------ ∥A∥∥B∥ - Find the Angle: To get θ by itself, you use the inverse cosine (acos).
/ A⋅B \ θ = arccos( ------ ) \ ∥A∥∥B∥ /
Visualizing the "Normalized" Shortcut
The text mentions: θ=cos(Â⋅B̂). (The "hat" symbol  means it is a normalized unit vector).
This is the "pro-gamer move" in graphics programming. If you normalize both vectors first (meaning you force their lengths to be 1), then ∥A∥ and ∥B∥ both become 1. The denominator in our formula disappears!
- If lengths are 1: The dot product is the cosine of the angle.
- To get the angle: You just plug that dot product into
acos().
A Practical Example: "Field of View"
Imagine you are coding a stealth game. You have a Guard and a Player.
- Vector A: The direction the Guard is looking.
- Vector B: The direction from the Guard to the Player.
If you normalize both vectors and calculate (A⋅B), you get the exact angle (e.g., 30°). If the Guard's peripheral vision is only 45°, you now know the Guard can see the player!
Important "Gotcha" for Programmers
When you use
acos()in languages like C#, C++, or Python, the result is usually in radians, not degrees.- Dot Product = 1: Angle is 0 (Vectors point same way).
- Dot Product = 0: Angle is 1.57 radians (90°).
- Dot Product = -1: Angle is 3.14 radians (180°).
Lee:
You said
If you normalize both vectors and calculate $acos(A⋅B), you get the exact angle (e.g., 30°). If the Guard's peripheral vision is only 45°, you now know the Guard can see the player!By "only 45°", is that from left to right of the guard's visibility? So, from straight ahead to the far right of his vision would only be 22.5°, thus the player is not visible at a 30° again from the guard? Or is that 45° from straight ahead to the far right of the guard's vision (for a total viewable angle of 90°)?Gemini:<br /> That is a fantastic catch. In game development and 3D graphics, this distinction is a common source of bugs!
When you use the dot product to check visibility, the angle θ you calculate is the offset from the center.
1. The "Half-Angle" Concept
The dot product measures the angle between two specific lines: where the guard is looking (the "forward vector") and where the player is. It doesn't know about "left" or "right"; it only knows "how far away from my nose is this object?"
- If your calculation acos(A⋅B) returns 30°, it means the player is 30° away from the center line.
- If the guard has a 45° Field of View (FOV), usually that refers to the total horizontal arc (22.5° to the left and 22.5° to the right).
In your specific scenario:
- Total FOV: 45°
- Allowed "Half-Angle": 22.5°
- Player's Angle: 30°
- Result: Not Visible. The player is "outside the cone."
2. How it's usually coded
To avoid doing the math every single frame, programmers usually define the guard's "View Cone" as a threshold value.
If the guard can see 45° to the left and 45° to the right (a 90° total FOV), the threshold is:<br />
cos(45°) ≈ 0.707The code then looks like this:```csharp // If dot product is greater than 0.707, the player is within the 90-degree cone if (DotProduct(GuardForward, DirectionToPlayer) > 0.707) { // I see you! }
```
3. Why the Dot Product is "Circular"
One interesting thing to visualize: because the dot product only cares about the angle between two vectors, the "visibility" it creates is actually a cone, not a flat triangle.
If you say "anything within 30° of my forward vector is visible," that includes 30° up, 30° down, 30° left, and 30° right. It creates a circular spotlight of vision.
Summary Table: FOV vs. Dot Product
| Total FOV | Angle from Center | Dot Product Threshold (Approx) | | ------------------ | ----------------- | ------------------------------ | | 180° (Flat wall) | 90° | 0.0 | | 90° (Standard) | 45° | 0.707 | | 60° (Narrow) | 30° | 0.866 | | 2° (Sniper Scope)| 1° | 0.999 | - Isolate the Cosine: Divide both sides by the magnitudes ().
-
-
blog.johanneslink.net blog.johanneslink.net
-
To Gen or Not To Gen: The Ethical Use of Generative AI 33 minute read This blog entry started out as a translation of an article that my colleague Jakob and I wrote for a German magazine. After that we added more stuff and enriched it by additional references and sources. We aim at giving an overview about many - but not all - aspects that we learned about GenAI and that we consider relevant for an informed ethical opinion. As for the depth of information, we are just scratching the surface; hopefully, the loads of references can lead you to diving in deeper wherever you want. Since we are both software developers our views are biased and distorted. Keep also in mind that any writing about a “hot” topic like this is nothing but a snapshot of what we think to know today. By the time you read it the authors’ knowledge and opinions have already changed. Last Update: December 8, 2025. Table of ContentsPermalink Abstract About us Johannes Link Jakob Schnell Introduction Ethics, what does that even mean? Clarification of terms Basics Can LLMs think? What LLMs are good at GenAI as a knowledge source GenAI in software development Actual vs. promised benefits Harmful aspects of GenAI GenAI is an ecological disaster Power Water Electronic Waste GenAI threatens education and science GenAI is destroying the free internet. GenAI is a danger to democracy GenAI versus human creativity Digital colonialism Political aspects Conclusion Can there be ethical GenAI? How to act ethically AbstractPermalink ChatGPT, Gemini, Copilot. The number of generative AI applications (GenAI) and models is growing every day. In the field of software development in particular, code generation, coding assistants and vibe coding are on everyone’s lips. Like any technology, GenAI has two sides. The great promises are offset by numerous disadvantages: immense energy consumption, mountains of electronic waste, the proliferation of misinformation on the internet and the dubious handling of intellectual property are just a few of the many negative aspects. Ethically responsible behaviour requires us to look at all the advantages, disadvantages and collateral damages of a technology before we use it or recommend its use to others. In this article, we examine both sides and eventually arrive at our personal and naturally subjective answer to whether and how GenAI can be used in an ethical manner. About usPermalink Johannes LinkPermalink … has been programming for over 40 years, 30 of them professionally. Since the end of the last century, extreme programming and other human-centred software development approaches have been at the heart of his work. The meaningful and ethical implementation of his private and professional life has been his driving force for years. He has been involved with GenAI since the early days of OpenAI’s GPT language models. More about Johannes can be found at https://johanneslink.net. Jakob SchnellPermalink … studied mathematics and computer science and has been working as a software developer for 5 years. He works as a lecturer and course director in university and non-university settings. As a youth leader, he also comes into regular contact with the lives of children and young people. In all these environments, he observes the growing use of GenAI and its impact on people. IntroductionPermalink Ethics, what does that even mean?Permalink Ethical behaviour sounds like the title of a boring university seminar. However, if you look at the wikipedia article of the term 1, you will find that ‘how individuals behave when confronted with ethical dilemmas’ is at the heart of the definition. So it’s about us as humans taking responsibility and weighing up whether and how we do or don’t do certain things based on our values. We have to consider ethical questions in our work because all the technologies we use and promote have an impact on us and on others. Therefore, they are neither neutral nor without alternative. It is about weighing up the advantages and potential against the damage and risks; and that applies to everyone, not just us personally. Because often those who benefit from a development are different from those who suffer the consequences. As individuals and as a society, we have the right to decide whether and how we want to use technologies. Ideally, this should be in a way that benefits us all; but under no circumstances should it be in a way that benefits a small group and harms the majority. The crux of the matter is that ethical behaviour does not come for free. Ethics are neither efficient nor do they enhance your economic profit. That means that by acting according to your values you will, at some point, have to give something up. If you’re not willing to do that, you don’t have values - just opinions. Clarification of termsPermalink When we write ‘generative AI’ (GenAI), we are referring to a very specific subset of the many techniques and approaches that fall under the term ‘artificial intelligence’. Strictly speaking, these are a variety of very different approaches that range from symbolic logic, over automated planning up to the broad field of machine learning (ML). Nowadays most effort, hype and money goes into deep learning (DL): a subfield of ML that uses multi-layered artificial neural networks to discover statistical correlations (aka patterns) based on very large amounts of training data in order to reproduce those patterns later. Large language models (LLM) and related methods for generating images, videos and speech now make it possible to apply this idea to completely unstructured data. While traditional ML methods often managed with a few dozen parameters, these models now work with several trillion (10^12) parameters. In order for this to produce the desired results, both the amount of training data and the training duration must be increased by several orders of magnitude. This brings us to the definition of what we mean by ‘GenAI’ in this article: Hyperscaled models that can only be developed, trained and deployed by a handful of companies in the world. These are primarily the GenAI services provided by OpenAI, Anthropic, Google and Microsoft, or based on these services. We also focus primarily on language models; the generation of images, videos, speech and music plays only a minor role in this article. Our focus on hyperscale services does not mean that other ML methods are free of ethical problems; however, we are dealing with a completely different order of magnitude of damage and risk here. For example, there do exist variations of GenAI that use the same or similar techniques, but on a much smaller scale and restricted domains (e.g. AlphaFold 2). These approaches tend to bring more value with fewer downsides. BasicsPermalink GenAI models are designed to interpolate and extrapolate 3, i.e. to fill in the gaps between training data and speculate beyond the limits of the training data. Together with the stochastic nature of the training data, this results in some interesting properties: GenAI models ‘invent’ answers; with LLMs, we like to refer to this as ‘hallucinations’. GenAI models do not know what is true or false, good or bad, efficient or effective, only what is statistically probable or improbable in relation to training data, context and query (aka prompt). GenAI models cannot explain their output; they have no capability of introspection. What is sold as introspection is just more output, with the previous output re-injected. GenAI models do not learn from you; they only draw from their training material. The learning experience is faked by reinjecting prior input into a conversation’s context 4. The context, i.e. the set of input parameters provided, is decisive for the accuracy of the generated result, but can also steer the model in the wrong direction. Increasing the context window makes a query much more computation-intensive - likely in a quadratic way. Therefore, the promised increase of “maximum context window” of many models is mostly fake 5. The reliability of LLMs cannot be fundamentally increased by even greater scaling 6. Can LLMs think?Permalink Proponents of the language-of-thought hypothesis 7 believe it is possible for purely language-based models to acquire the capabilities of the human brain – reasoning, modelling, abstraction and much more. Some enthusiasts even claim that today’s models have already acquired this capability. However, recent studies 8 9 show that today’s models are neither capable of genuine reasoning nor do they build internal models of the world. Moreover, “…according to current neuroscience, human thinking is largely independent of human language 10” and there is fundamental scientific doubt that achieving human cognition through computation is achievable in practice let alone by scaling up training of deep networks 11. An example of a lack of understanding of the world is the prompt ‘Give me a random number between 0 and 50’. The typical GenAI response to this is ‘27’, and it is significantly more reliable than true randomness would allow. (If you don’t believe it, just try it out!) This is because 27 is the most likely answer in the GenAI training data – and not because the model understands what ‘random’ means. ‘Chain of Thought (CoT)’ approaches and ‘Reasoning models’ attempt to improve reasoning by breaking down a prompt, the query to the model, into individual (logical) steps and then delegating these individual steps back to the LLM. This allows some well-known reasoning benchmarks to be met, but it also multiplies the necessary computational effort by a factor between 30 and 700 12. In addition, multistep reasoning lets individual errors chain together to form large errors. And yet, CoT models do not seem to possess any real reasoning abilities 13 14 and improve the overall accuracy of LLMs only marginally 15. The following thought experiment from 16 underscores the lack of real “thinking” capabilities: LLMs have simultaneous access to significantly more knowledge than humans. Together with the postulated ability of LLMs to think logically and draw conclusions, new insights should just fall from the sky. But they don’t. Getting new insights from LLMs would require these to be already encoded in the existing training material, and to be decoded and extracted by pure statistical means. What LLMs are good atPermalink Undoubtedly, LLMs represent a major qualitative advance when it comes to extracting information from texts, generating texts in natural and artificial languages, and machine translation. But even here, the error rate, and above all the type of error (‘hallucinations’), is so high that autonomous, unsupervised use in serious applications must be considered highly negligent. GenAI as a knowledge sourcePermalink As we have pointed out above, LLMs cannot differentiate between true and false - regardless of the training material. It does not answer the question “What is XYZ?” but the question “How would an answer to question ‘What is XYZ?’ look like?”. Nevertheless, many people claim that the answers that ChatGPT and alike provide for the typical what-how-when-who queries are good enough and often better than what a “normal” web search would have given us. Arguably, this is the most prevalent use case for “AI” bots today. The problem is that most of the time we will never learn about the inaccuracies, left-outs, distortions and biases that the answer contained - unless we re-check everything, which defies the whole purpose of speeding up knowledge retrieval. The less we already know, the better the “AI’s” answer looks to us, but the less equipped we are to spot the problems. A recent by the BBC and 22 Public Service Media organizations shows that 45% of all “AI” assistants’ answers on questions about news and current affairs have significant errors 17. Moreover, LLMs are easy prey for manipulation - either by the service providing organization or by third parties. A recent study claims that even multi-billion-parameter models can be “poisoned” by injecting just a few corrupted documents 18. So, if anything is at stake all output from LLMs must be carefully validated. Doing that, however, would contradict the whole point of using “AI” to speed up knowledge acquisition. GenAI in software developmentPermalink The creation and modification of computer programmes is considered a prime domain for the use of LLMs. This is partly because programming languages have less linguistic variance and ambiguity than natural languages. Moreover, there are many methods for automatically checking generated source code, such as compiling, static code analysis and automated testing. This simplifies the validation of generated code and thereby gives an additional feeling of trust. Nevertheless, individual reports on the success of coding assistants such as Copilot, Cursor, etc. vary greatly. They range from ‘completely replacing me as a developer’ to ‘significantly hindering my work’. Some argue that coding agents considerably reduce the time they have to invest in “boilerplate” work, like writing tests, creating data transfer objects or connecting your domain code to external libraries. Others counter by pointing out that delegating these drudgeries to GenAI makes you miss opportunities to get rid of them, e.g. by introducing a new abstraction or automating parts of your pipeline, and to learn about the intricacies and failure modes of the external library. Other than old-school code generation or code libraries prompting a coding agent is not “just another layer of abstraction”. It misses out on several crucial aspects of a useful abstraction: Its output is not deterministic. You cannot rely on any agent producing the same code next time you feed it the same prompt. The agent does not hide the implementation details, nor does it allow you to reliably change those details if the previous implementation turns out to be inadequate. Code that is output by an LLM, even if it is generated “for free”, has to be considered and maintained each time you touch the related logic or feature. The agent does not tell you if the amount of details you give in your prompt is sufficient for figuring out an adequate implementation. On the contrary, the LLM will always fill the specification holes with some statistically derived assumptions. Sadly, serious studies on the actual benefits of GenAI in software development are rare. The randomised trial by Metr 19 provides an initial indication, measuring a decline in development speed for experienced developers. An informal study by ThoughtWorks estimates the potential productivity gain from using GenAI in software development at around 5-15% 20. If “AI coding” were increasing programmers’ productivity by any big number, we would see a measurable growth of new software in app stores and OSS repositories. But we don’t, the numbers are flat at best 2122. But even if we assume a productivity increase in coding through GenAI, there are still two points that further diminish this postulated efficiency gain: Firstly, the results of the generation must still be cross-checked by human developers. However, it is well known that humans are poor checkers and lose both attention and enjoyment in the process. Secondly, software development is only to a small extent about writing and changing code. The most important part is discovering solutions and learning about the use of these solutions in their context. Peter Naur calls this ‘programming as theory building’ 23. Even the perfect coding assistant can therefore only take over the coding part of software development. For the essential rest, we still need humans. If we now also consider the finding that using AI can relatively quickly lead to a loss of problem-solving skills 24 or that these skills are not acquired at all, then the overall benefit of using GenAI in professional software development is more than questionable. As long as programming - and every technicality that comes with it - will not be fully replaced by some kind of AI, we will still need expert developers who can programm, maintain and debug code to the finest level of detail. Where, we wonder, will those senior developers come from when companies replace their junior staff with coding agents? Actual vs. promised benefitsPermalink If you read testimonials about the use of GenAI that people perceive as successful, you will mostly encounter scenarios in which ‘AI’ helps to make tasks that are perceived as boring, unnecessarily time-consuming or actually pointless faster or more pleasant. So it’s mainly about personal convenience and perceived efficiency. Entertainment also plays a major role: the poem for Grandma’s birthday, the funny song for the company anniversary or the humorous image for the presentation are quickly and supposedly inexpensively generated by ‘AI’. However, the promises made by the dominant GenAI companies are quite different: solving the climate crisis, providing the best medical advice for everyone, revolutionising science, ‘democratising’ education and much more. GPT5, for example, is touted by Sam Altman, CEO of OpenAI, as follows: ‘With GPT-5, it’s now like talking to an expert — a legitimate PhD-level expert in any area you need […] they can help you with whatever your goals are.’ 25 However, to date, there is still no actual use case that provides a real qualitative benefit for humanity or at least larger groups. The question ‘What significant problem (for us as a society) does GenAI solve?’ remains unanswered. On the contrary: While machine learning and deep learning methods certainly have useful applications, the most profitable area of application for ‘AI’ at present is the discovery and development of new oil and gas fields 26. Harmful aspects of GenAIPermalink But regardless of how one assesses the benefits of this technology, we must also consider the downsides, because only then can we ultimately make an informed and fair assessment. In fact, the range of negative effects of hyperscaled generative AI that can already be observed is vast. Added to this are numerous risks that have the potential to cause great social harm. Let’s take a look at what we consider to be the biggest threats: GenAI is an ecological disasterPermalink PowerPermalink The data centres required for training and operating large generative models 27 far exceed today’s dimensions in terms of both number and size. The projected data centre energy demand in the USA is predicted to grow from 4.4% of total electricity in 2023 to 22% in 2028 28. In addition, the typical data centre electricity mix is more CO2-intensive than the average mix. There is an estimated raise of ~11 percent for coal generated electricity in the US, as well as tripled emissions of greenhouse gases worldwide by 2030 - compared to the scenario without GenAI technology 29. Just recently Sam Altman from OpenAI blogged some numbers about the energy and water usage of ChatGPT for “the average query” 30. On the one hand, an average is rather meaningless when a distribution is heavily unsymmetric; the numbers for queries with large contexts or “chain of reasoning” computations would be orders of magnitude higher. Thus, the potential efficiency gains from more economical language models are more than offset by the proliferation of use, e.g. through CoT approaches and ‘agent systems’. On the other hand, big tech’s disclosure of energy consumption (e.g. by Google 31) is intentionally selective. Ketan Joshi goes into quite some details why experts think that the AI industry is hiding the full picture 32. Since building new power plants - even coal or gas fuelled ones - takes a lot of time, data center companies are even reviving old jet engines for powering their new hyper-scalers 33. You have to be aware that those engines are not only much more noisy than other power plants but also pump out nitrous oxide, one of the main chemicals responsible for acid rain 34. WaterPermalink Another problem is the immensely high water consumption of these data centres 35. After all, cooling requires clean water in drinking quality in order to not contaminate or clog the cooling pipes and pumps. Already today, new data centre locations are competing with human consumption of drinking water. According to Bloomberg News about two-thirds of data-centers that were built or developed in 2022 are located in areas that are already under “water-stress” 36. In the US alone “AI servers […] could generate an annual water footprint ranging from 731 to 1,125 million m3” 37. It’s not only an American problem, though. In other areas of the world the water-thirsty data centers also compete with the drinking water supply for humans 38. Electronic WastePermalink Another ecological problem is being noticeably exacerbated by ‘AI’: the amount of electronic waste (e-waste) that we ship mainly to “Third World” countries and which is responsible for soil contamination there. Efficient training and querying of very large neural networks requires very large quantities of specialised chips (GPUs). These chips often have to be replaced and disposed of within two years. The typical data center might not last longer than 3 to 5 years before it has to be rebuilt in large parts39. In summary, it can be said that GenAI is at least an accelerator of the ecological catastrophe that threatens the earth. And it is the argument for Google, Amazon and Microsoft to completely abolish their zero CO2 targets 40 and replace them with investments of several hundred billion dollars for new data centers. GenAI threatens education and sciencePermalink People often try to use GenAI in areas where they feel overloaded and overwhelmed: training, studying, nursing, psychotherapeutic care, etc. The fields of application for ‘AI’ are therefore a good indication of socially neglected and underfunded areas. The fact that LLMs are very good at conveying the impression of genuine knowledge and competence makes their use particularly attractive in these areas. A teacher under the simultaneous pressure of lesson preparation, corrections and covering for sick colleagues turns to ChatGPT to quickly create an exercise sheet. A student under pressure to get good grades has their English essay corrected by ‘AI’. The researcher under pressure to publish will ‘save’ research time by reading the AI-generated summary of relevant papers – even if they are completely wrong in terms of content 41. Tech companies like OpenAI and Microsoft play on that situation by offering their ‘AI’ for free or for little money to students and universities. The goal is obvious: Students that get hooked on outsourcing some of their “tedious” task to a service will continue to use - and eventually buy - this service after graduation. What falls by the wayside are problem-solving skills, engagement with complex sources, and the generation of knowledge through understanding and supplementing existing knowledge. Some even argue that AI is destroying critical education and learning itself 42: Students aren’t just learning less; their brains are learning not to learn. The training cycle of schools and universities is fast. Teachers are already reporting that pupils and students have acquired noticeably less competence in recent years, but have instead become dependent on unreliable ‘tools’ 43. The real problem with using GenAI to do assignments is not cheating, but students “are not just undermining their ability to learn, but to someday lead.” 44 GenAI is destroying the free internet.Permalink The fight against bots on the internet is almost as old as the internet itself – and has been quite successful so far. Multifactor authentication, reCaptcha, honeypots and browser fingerprinting are just a few of the tools that help protect against automated abuse. However, GenAI takes this problem to a new level – in two ways. To make ‘the internet’ usable as the main source for training LLMs, AI companies use so-called ‘crawlers’. These essentially behave like DDoS attackers: They send tens of thousands of requests at once, from several hundred IPs in a very short time. Robot.txt files are ignored; instead, the source IP and user agent are obscured 45. These practices have massive disadvantages for providers of genuine content: Costs for additional bandwidth. Lost advertising revenue, as search engines now offer LLM-generated summaries instead of links to the sources. This threatens the existence of remaining independent journalism in particular 46. Misuse of own content for AI-supported competition. If the place where knowledge is generated is separated from the place where it is consumed, and if this makes the performance of generation even more opaque than before, the motivation to continue generating knowledge also declines. For projects such as Wikipedia, this means fewer donors and fewer contributors. Open communities often have no other option but to shut themselves off. Another aspect is the flooding of the internet with generated content that cannot be automatically distinguished from non-generated content. This content overwhelms the maintainers of open source software or portals such as Wikipedia 47. If this content is then also entered by humans – often in the belief that they are doing good – it is no longer possible to take action against the methodology. In the long run, this means that less and less authentic training material will lead to increasingly poor results from the models. Last but not least, autonomously acting agents make the already dire state of internet security much worse 48. Think of handing all your personal data and credentials to a robot that is distributing and using that data across the web, wherever and whenever it deems it necessary for reaching some goal. is controlled by LLMs who are vulnerable to all kinds of prompt injection attacs 49. is controlled by and reporting to companies that do not have your best interest in mind. has no awareness and knowledge about the implication of its actions. is acting on your behalf and thereby making you accountable. GenAI is a danger to democracyPermalink The manipulation of public opinion through social media precedes the arrival of LLMs. However, this technology gives the manipulators much more leverage. By flooding the web with fake news, fake videos and fake everything undemocratic (or just criminal) parties make it harder and harder for any serious media and journalism to get the attention of the public. People no longer have a common factual basis, which is necessary for all social negotiations. If you don’t agree on at least some basic facts, arguing about policies and measures to take is pointless. Without negotiations democracy will be dying; in many parts of the world it already is. GenAI versus human creativityPermalink Art and creativity are also threatened by generative AI. The impact on artists’ incomes of logos, images and illustrations now being easily and quickly created by AI prompts is obvious. A similar effect can also be observed in other areas. Studies show that poems written by LLMs are indistinguishable from those written by humans and that generative AI products are often rated more highly 50. This can be explained by a trend towards the middle and the average, which can also be observed in the music and film scenes film scene: due to its basic function, GenAI cannot create anything fundamentally new, but replicates familiar patterns, which is precisely why it is so well received by the public. Ironically, ‘AI’ draws its ‘creativity’ from the content of those it seeks to replace. Much of this content was used as training material against the will of the rights holders. Whether this constitutes a copyright infringement has not yet been decided; morally, the situation seems clear. The creative community is the first to be seriously threatened by GenAI in its livelihood 51. It’s not a coincidence that a big part of GenAI efforts is targeted at “democratizing art”. This framing is completely upside down. Art has been one of the most democratic activities for a very long time. Everybody can do it; but not everybody wants to do put in the effort, the practicing time and the soul. Real art is not about the product but about the process, which requires real humans. Generating art without the friction is about getting rid of the humans in the loop - and still making money. Digital colonialismPermalink The huge amount of data required by hyperscaled AI approaches makes it impossible to completely curate the learning content. And yet, one would like to avoid the reproduction of racist, inhuman and criminal content. Attempts are being made to get the problem under control by subsequently adapting the models to human preferences and local laws through additional ‘reinforcement learning from human feedback (RLHF)’ 52. The cheap labour for this very costly process can be found in the Global South. There, people are exposed to hours of hate speech, child abuse, domestic violence and other horrific scenarios in their poorly paid jobs in order to filter them out of the training material of large AI companies 53. Many emerge from these activities traumatised. However, it is not only people who are exploited in the less developed regions of the world, but also nature: the poisoning of the soil with chemicals during the extraction of raw materials for digital chips, as well as the contamination caused by our electronic waste and its improper disposal, are collateral damage that we willingly accept and whose long-term consequences are currently extremely difficult to assess. Here, too, the “developed” world profits, whereas the negative aspects are outsourced to the former colonies and other poor regions of the world. Political aspectsPermalink As software developers, we would like to ‘leave politics out of it’ and instead focus entirely on the cool tech. However, this is impossible when the advocates of this technology pursue strong political and ideological goals. In the case of GenAI, we can cleary see that the US corporations behind it (OpenAI, Google, Meta, Microsoft, etc.) have no problem with the current authoritarian – some say fascist – US government 54. In concrete terms, this means, among other things, that the models are explicitly manipulated to be less liberal or simply not to generate any output that could upset the CEO or the president 55. Even more serious is the fact that many of the leading minds behind these corporations and their financiers adhere to beliefs that can be broadly described as digital fascism. These include Peter Thiel, Marc Andreessen, Alex Karp, JD Vance, Elon Musk and many others on “The Authoritarian Stack” 56. Their ideologies, disguised as rational theories, are called longtermism and effective altruism. What they have in common is that they consider democracy and the state to be obsolete models, compassion to be ‘woke’, and that the current problems of humanity are insignificant, as our future lies in the colonisation of space and the merging of humans with artificial superintelligence 57. Do we want to give people who adhere to these ideologies (even) more power, money and influence by using and paying for their products? Do we want to feed their computer systems with our data? Do we really want to expose ourselves and our children to the answers from chatbots which they have manipulated? Not quite as abstruse, but similarly misanthropic, is the imminent displacement of many jobs by AI, as postulated by the same corporations in order to put pressure on employees with this claim. Demanding a large salary? Insisting on your legal rights? Complaining about too much workload? Doubts about the company’s goals? Then we’ll just replace you with cheap and uncomplaining AI! Whichever way you look at it, AI and GenAI are already being used politically. If we go along without resistance, we are endorsing this approach and supporting it with our time, our attention and our money. ConclusionPermalink Ideally, we would like to quantify our assessment by adding up the advantages, adding up the disadvantages and finally checking whether the balance is positive or negative. Unfortunately, in our specific case, neither the benefits nor the harm are easily quantifiable; we must therefore consult our social and personal values. Discussions about GenAI usually revolve purely around its benefits. Often, the capabilities of all ‘AI’ technologies (e.g. protein folding with AlphaFold 2) are lumped together, even though they have little in common with hyperscaling GenAI. However, if we consider the consequences and do not ignore the problems this technology entails – i.e. if we consider both sides in terms of ethics – the assessment changes. Convenience, speed and entertainment are then weighed against numerous damages and risks to the environment, the state and humanity. In this sense, the ethical use and further expansion of GenAI in its current form is not possible. Can there be ethical GenAI?Permalink If the use of GenAI is not ethical today what would have to change, which negative effects of GenAI would have to disappear or at least be greatly reduced in order to tip the balance between benefits and harms in the other direction? The models would have to be trained exclusively with publicly known content whose original creators consent to its use in training AI models. The environmental damage would have to be reduced to such an extent that it does not further fuel the climate crisis. Society would have to get full access to the training and operation of the models in order to rule out manipulation by third parties and restrict their use to beneficial purposes. This would require democratic processes, good regulation and oversight through judges and courts. The misuse and harming of others, e.g., through copyright theft or digital colonialism, would have to be prevented. Is such a change conceivable? Perhaps. Is it likely, given the interest groups and political aspects involved? Probably not
All these factors are achievable I think, or will be soonish. Smaller models, better sourced data sets, niche models, etc. But not with current actors as mentioned at the end.
-
-
pierce.dev pierce.dev
-
I'm not advocating that everyone should self-host everything. But the pendulum has swung too far toward managed services. There's a large sweet spot where self-hosting makes perfect sense, and more teams should seriously consider it. Start small. If you're paying more than $200/month for RDS, spin up a test server and migrate a non-critical database. You might be surprised by how straightforward it is. The future of infrastructure is almost certainly more hybrid than it's been recently: managed services where they add genuine value, self-hosted where they're just expensive abstractions. Postgres often falls into the latter category. Footnotes They're either just hosting a vanilla postgres instance that's tied to the deployed hardware config, or doing something opaque with edge deploys and sharding. In the latter case they near guarantee your DB will stay highly available but costs can quickly spiral out of control. ↩ Maybe up to billions at this point. ↩ Even on otherwise absolutely snail speed hardware. ↩ This was Jeff Bezos's favorite phrase during the early AWS days, and it stuck. ↩ Similar options include OVH, Hetzner dedicated instances, or even bare metal from providers like Equinix. ↩ AWS RDS & S3 has had several major outages over the years. The most memorable was the 2017 US-East-1 outage that took down half the internet. ↩
Cloud hosting can become an expensive abstraction layer quickly. I also think there's an entire generation of coders / engineers who treat silo'd cloudhosting as a given, without considering other options and their benefits. Large window for selfhosting in which postgres almost always falls
-
When self-hosting doesn't make sense I'd argue self-hosting is the right choice for basically everyone, with the few exceptions at both ends of the extreme: If you're just starting out in software & want to get something working quickly with vibe coding, it's easier to treat Postgres as just another remote API that you can call from your single deployed app If you're a really big company and are reaching the scale where you need trained database engineers to just work on your stack, you might get economies of scale by just outsourcing that work to a cloud company that has guaranteed talent in that area. The second full freight salaries come into play, outsourcing looks a bit cheaper. Regulated workloads (PCI-DSS, FedRAMP, HIPAA, etc.) sometimes require a managed platform with signed BAAs or explicit compliance attestations.
Sees use for silo'd postgres hosting on the extremes of the spectrum: when you start without knowledge and are vibecoding, so you can treat the database as just another API, and when you are megacorp (outsourcing looks cheaper quickly if you have to otherwise pay multiple FTE salaries otherwise), or/and have to prove regulatory compliance.
-
For the most part managed database services aren't running some magical proprietary technology. They're just running the same open-source Postgres you can download with some operational tooling wrapped around it. Take AWS RDS. Under the hood, it's: Standard Postgres compiled with some AWS-specific monitoring hooks A custom backup system using EBS snapshots Automated configuration management via Chef/Puppet/Ansible Load balancers and connection pooling (PgBouncer) Monitoring integration with CloudWatch Automated failover scripting
AWS RDS is not much else as open source postgres with operational tooling that is not complex itself.
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the original reviews.
Public Reviews:
Reviewer #1 (Public review):
Summary:
Zhang and colleagues examine neural representations underlying abstract navigation in the entorhinal cortex (EC) and hippocampus (HC) using fMRI. This paper replicates a previously identified hexagonal modulation of abstract navigation vectors in abstract space in EC in a novel task involving navigating in a conceptual Greeble space. In HC, the authors claim to identify a three-fold signal of the navigation angle. They also use a novel analysis technique (spectral analysis) to look at spatial patterns in these two areas and identify phase coupling between HC and EC. Finally, the authors propose an EC-HPC PhaseSync Model to understand how the EC and HC construct cognitive maps. While the wide array of techniques used is impressive and their creativity in analysis is admirable, overall, I found the paper a bit confusing and unconvincing. I recommend a significant rewrite of their paper to motivate their methods and clarify what they actually did and why. The claim of three-fold modulation in HC, while potentially highly interesting to the community, needs more background to motivate why they did the analysis in the first place, more interpretation as to why this would emerge in biology, and more care taken to consider alternative hypotheses seeped in existing models of HC function. I think this paper does have potential to be interesting and impactful, but I would like to see these issues improved first.
General comments:
(1) Some of the terminology used does not match the terminology used in previous relevant literature (e.g., sinusoidal analysis, 1D directional domain).
We thank the reviewer for this valuable suggestion, which helps to improve the consistency of our terminology with previous literature and to reduce potential ambiguity. Accordingly, we have replaced “sinusoidal analysis” with “sinusoidal modulation” (Doeller et al., 2010; Bao et al., 2019; Raithel et al., 2023) and “1D directional domain” with “angular domain of path directions” throughout the manuscript.
(2) Throughout the paper, novel methods and ideas are introduced without adequate explanation (e.g., the spectral analysis and three-fold periodicity of HC).
We thank the reviewer for raising this important point. In the revised manuscript, we have substantially extended the Introduction (paragraphs 2–4) to clarify our hypothesis, explicitly explaining why the three primary axes of the hexagonal grid cell code may manifest as vector fields. We have also revised the first paragraph of the “3-fold periodicity in the HPC” section in the Results to clarify the rationale for using spectral analysis. Please refer to our responses to comment 2 and 3 below for details.
Reviewer #2 (Public review):
The authors report results from behavioral data, fMRI recordings, and computer simulations during a conceptual navigation task. They report 3-fold symmetry in behavioral and simulated model performance, 3-fold symmetry in hippocampal activity, and 6-fold symmetry in entorhinal activity (all as a function of movement directions in conceptual space). The analyses are thoroughly done, and the results and simulations are very interesting.
We sincerely thank the reviewer for the positive and encouraging comments on our study.
Recommendations for the authors:
Reviewer #1 (Recommendations for the authors):
(1) This paper has quite a few spelling and grammatical mistakes, making it difficult to understand at times.
We apologize for the wordings and grammatical errors. We have thoroughly re-read and carefully edited the entire manuscript to correct typographical and grammatical errors, ensuring improved clarity and readability.
(2) Introduction - It's not clear why the three primary axes of hexagonal grid cell code would manifest as vector fields.
We thank the reviewer for raising this important point. In the revised Introduction (paragraphs 2, 3, and 4), we now explicitly explain the rationale behind our hypothesis that the three primary axes of the hexagonal grid cell code manifest as vector fields.
In paragraph 2, we present empirical evidence from rodent, bat, and human studies demonstrating that mental simulation of prospective paths relies on vectorial representations in the hippocampus (Sarel et al., 2017; Ormond and O’Keefe, 2022; Muhle-Karbe et al., 2023).
In paragraphs 3 and 4, we introduce our central hypothesis: vectorial representations may originate from population-level projections of entorhinal grid cell activity, based on three key considerations:
(1) The EC serves as the major source of hippocampal input (Witter and Amaral, 1991; van Groen et al., 2003; Garcia and Buffalo, 2020).
(2) Grid codes exhibit nearly invariant spatial orientations (Hafting et al., 2005; Gardner et al., 2022), which makes it plausible that their spatially periodic activity can be detected using fMRI.
(3) A model-based inference: for example, in the simplest case, when one mentally simulates a straight pathway aligned with the grid orientation, a subpopulation of grid cells would be activated. The resulting population activity would form a near-perfect vectorial representation, with constant activation strength along the path. In contrast, if the simulated path is misaligned with the grid orientation, the population response becomes a distorted vectorial code. Consequently, simulating all possible straight paths spanning 0°–360° results in 3-fold periodicity in the activity patterns—due to the 180° rotational symmetry of the hexagonal grid, orientations separated by 180° are indistinguishable.
We therefore speculate that vectorial representations embedded in grid cell activity exhibit 3-fold periodicity across spatial orientations and serve as a periodic structure to represent spatial direction. Supporting this view, reorientation paradigms in both rodents and young children have shown that subjects search equally in two opposite directions, reflecting successful orientation encoding but a failure to integrate absolute spatial direction (Hermer and Spelke, 1994; Julian et al., 2015; Gallistel, 2017; Julian et al., 2018).
(3) It took me a few reads to understand what the spectral analysis was. After understanding, I do think this is quite clever. However, this paper needs more motivation to understand why you are performing this analysis. E.g., why not just take the average regressor at the 10º, 70º, etc. bins and compare it to the average regressor at 40º, 100º bins? What does the Fourier transform buy you?
We are sorry for the confusion. we outline the rationale for employing Fast Fourier Transform (FFT) analysis to identify neural periodicity. In the revised manuscript, we have added these clarifications into the first paragraph of the “3-fold periodicity in the HPC” subsection in the Results.
First, FFT serves as an independent approach to cross-validate the sinusoidal modulation results, providing complementary evidence for the 6-fold periodicity in EC and the 3-fold periodicity in HPC.
Second, FFT enables unbiased detection of multiple candidate periodicities (e.g., 3–7-fold) simultaneously without requiring prior assumptions about spatial phase (orientation). By contrast, directly comparing “aligned” versus “misaligned” angular bins (e.g., 10°/70° vs. 40°/100°) would implicitly assume knowledge of the phase offset, which was not known a priori.
Finally, FFT uniquely allows periodicity analysis of behavioral performance, which is not feasible with standard sinusoidal GLM approaches. This methodological consistency makes it possible to directly compare periodicities across neural and behavioral domains.
(4) A more minor point: at one point, you say it’s a spectral analysis of the BOLD signals, but the methods description makes it sound like you estimated regressors at each of the bins before performing FFT. Please clarify.
We apologize for the confusion. In our manuscript, we use the term spectral analysis to distinguish this approach from sinusoidal modulation analysis. Conceptually, our spectral analysis involves a three-level procedure:
(1) First level: We estimated direction-dependent activity maps using a general linear model (GLM), which included 36 regressors corresponding to path directions, down-sampled in 10° increments.
(2) Second level: We applied a Fast Fourier Transform (FFT) to the direction-dependent activity maps derived from the GLM to examine the spectral magnitude of potential spatial periodicities.
(3) Third level: We conducted group-level statistical analyses across participants to assess the consistency of the observed periodicities.
We have revised the “Spectral analysis of MRI BOLD signals” subsection in the Methods to clarify this multi-level procedure.
(5) Figure 4a:
Why do the phases go all the way to 2*pi if periodicity is either three-fold or six-fold?
When performing correlation between phases, you should perform a circular-circular correlation instead of a Pearson's correlation.
We thank the reviewer for raising this important point. In the original Figure 4a, both EC and HPC phases spanned 0–2π because their sinusoidal phase estimates were projected into a common angular space by scaling them according to their symmetry factors (i.e., multiplying the 3-fold phase by 3 and the 6-fold phase by 6), followed by taking the modulo 2π. However, this projection forced signals with distinct intrinsic periodicities (120° vs. 60° cycles) into a shared 360° space, thereby distorting their relative angular distances and disrupting the one-to-one correspondence between physical directions and phase values. Consequently, this transformation could bias the estimation of their phase relationship.
In the revised analysis and Figure 4a, we retained the original phase estimates derived from the sinusoidal modulation within their native periodic ranges (0–120° for 3-fold and 0–60° for 6-fold) by applying modulo operations directly. Following your suggestion, the relationship between EC and HPC phases was then quantified using circular–circular correlation (Jammalamadaka & Sengupta, 2001), as implemented in the CircStat MATLAB toolbox. This updated analysis avoids the rescaling artifact and provides a statistically stronger and conceptually clearer characterization of the phase correspondence between EC and HPC.
(6) Figure 4d needs additional clarification:
Phase-locking is typically used to describe data with a high temporal precision. I understand you adopted an EEG analysis technique to this reconstructed fMRI time-series data, but it should be described differently to avoid confusion. This needs additional control analyses (especially given that 3 is a multiple of 6) to confirm that this result is specific to the periodicities found in the paper.
We thank the reviewer for this insightful comment. We have extensively revised the description of the Figure 4 to avoid confusion with EEG-based phase-locking techniques. The revised text now explicitly clarifies that our approach quantifies spatial-domain periodic coupling across path directions, rather than temporal synchronization of neural signals.
To further address the reviewer’s concern about potential effects of the integer multiple relationship between the 3-fold HPC and 6-fold EC periodicities, we additionally performed two control analyses using the 9-fold and 12-fold EC components, both of which are also integer multiples of the 3-fold HPC periodicity. Neither control analysis showed significant coupling (p > 0.05), confirming that the observed 3-fold–6-fold coupling was specific and not driven by their harmonic relationship.
The description of the revised Figure 4 has been updated in the “Phase Synchronization Between HPC and EC Activity” subsection of the Results.
(7) Figure 5a is misleading. In the text, you say you test for propagation to egocentric cortical areas, but I don’t see any analyses done that test this. This feels more like a possible extension/future direction of your work that may be better placed in the discussion.
We are sorry for the confusion. Figure 5a was intended as a hypothesis-driven illustration to motivate our analysis of behavioral periodicity based on participants’ task performance. However, we agree with the reviewer that, on its own, Figure 5a could be misleading, as it does not directly present supporting analyses.
To provide empirical support for the interpretation depicted in Figure 5a, we conducted a whole-brain analysis (Figure S8), which revealed significant 3-fold periodic signals in egocentric cortical regions, including the parietal cortex (PC), precuneus (PCU), and motor regions.
To avoid potential misinterpretation, we have revised the main text to include these results and explicitly referenced Figure S8 in connection with Figure 5a.
The updated description in the “3-fold periodicity in human behavior” subsection in the Results is as follows:
“Considering the reciprocal connectivity between the medial temporal lobe (MTL), where the EC and HPC reside, and the parietal cortex implicated in visuospatial perception and action, together with the observed 3-fold periodicity within the DMN (including the PC and PCu; Fig. S8), we hypothesized that the 3-fold periodic representations of path directions extend beyond the MTL to the egocentric cortical areas, such as the PC, thereby influencing participants' visuospatial task performance (Fig. 5a)”.
Additionally, Figure 5a has been modified to more clearly highlight the hypothesized link between activity periodicity and behavioral periodicity, rather than suggesting a direct anatomical pathway.
(8) PhaseSync model: I am not an expert in this type of modeling, so please put a lower weight on this comment (especially compared to some of the other reviewers). While the PhaseSync model seems interesting, it’s not clear from the discussion how this compares to current models. E.g., Does it support them by adding the three-fold HC periodicity? Does it demonstrate that some of them can't be correct because they don't include this three-fold periodicity?
We thank the reviewer for the insightful comment regarding the PhaseSync model. We agree that further clarifying its relationship to existing computational frameworks is important.
The EC–HPC PhaseSync model is not intended to replace or contradict existing grid–place cell models of navigation (e.g., Bicanski and Burgess, 2019; Whittington et al., 2020; Edvardsen et al., 2020). Instead, it offers a hierarchical extension by proposing that vectorial representations in the hippocampus emerge from the projections of periodic grid codes in the entorhinal cortex. Specifically, the model suggests that grid cell populations encode integrated path information, forming a vectorial gradient toward goal locations.
To simplify the theoretical account, our model was implemented in an idealized square layout. In more complex real-world environments, hippocampal 3-fold periodicity may interact with additional spatial variables, such as distance, movement speed, and environmental boundaries.
We have revised the final two paragraphs of the Discussion to clarify this conceptual framework and emphasize the importance of future studies in exploring how periodic activity in the EC–HPC circuit interacts with environmental features to support navigation.
Reviewer #2 (Recommendations for the authors):
(1) Please show a histogram of movement direction sampling for each participant.
We thank the reviewer for this helpful suggestion. We have added a new supplementary figure (Figure S2) showing histograms of path direction sampling for each participant (36 bins of 10°). The figure is also included. Rayleigh tests for circular uniformity revealed no significant deviations from uniformity (all ps > 0.05, Bonferroni-corrected across participants), confirming that path directions were sampled evenly across 0°–360°.
(2) Why didn’t you use participants’ original trajectories (instead of the trajectories inferred from the movement start and end points) for the hexadirectional analyses?
In our paradigm, participants used two MRI-compatible 2-button response boxes (one for each hand) to adjust the two features of the greebles. As a result, the raw adjustment path contained only four cardinal directions (up, down, left, right). If we were to use the raw stepwise trajectories, the analysis would be restricted to these four directions, which would severely limit the angular resolution. By instead defining direction as the vector from the start to the end position in feature space, we can expand the effective range of directions to the full 0–360°. This approach follows previous literature on abstract grid-like coding in humans (e.g., Constantinescu et al., 2016), where direction was similarly defined by the relative change between two feature dimensions rather than the literal stepwise path. We have added this clarification in the “Sinusoidal modulation” subsection of the revised method.
(3) Legend of Figure 2: the statement "localizing grid cell activity" seems too strong because it is still not clear whether hexadirectional signals indeed result from grid-cell activity (e.g., Bin Khalid et al., eLife, 2024). I would suggest rephrasing this statement (here and elsewhere).
Thank you for this helpful suggestion. We have removed the statement “localizing grid cell activity” to avoid ambiguity and revised the legend of Figure 2a to more explicitly highlight its main purpose—defining how path directions and the aligned/misaligned conditions were constructed in the 6-fold modulation. We have also modified similar expressions throughout the manuscript to ensure consistency and clarity.
(4) Legend of Figure 2: “cluster-based SVC correction for multiple comparisons” - what is the small volume you are using for the correction? Bilateral EC?
For both Figure 2 and Figure 3, the anatomical mask of the bilateral medial temporal lobe (MTL), as defined by the AAL atlas, was used as the small volume for correction. This has been clarified in the revised Statistical Analysis section of the Methods as “… with small-volume correction (SVC) applied within the bilateral MTL”.
(5) Legend of Figure 2: "ROI-based analysis" - what kind of ROI are you using? "corrected for multiple comparisons" - which comparisons are you referring to? Different symmetries and also the right/left hemisphere?
In Figure 2b, the ROI was defined as a functional mask derived from the significant activation cluster in the right entorhinal cortex (EC). Since no robust clusters were observed in the left EC, the functional ROI was restricted to the right hemisphere. We indeed included Figure 2c to illustrate this point; however, we recognize that our description in the text was not sufficiently clear.
Regarding the correction for multiple comparisons, this refers specifically to the comparisons across different rotational symmetries (3-, 4-, 5-, 6-, and 7-fold). Only the 6-fold symmetry survived correction, whereas no significant effects were detected for the other symmetries.
We have clarified these points in the “6-fold periodicity in the EC” subsection of the result as “… The ROI was defined as a functional mask of the right EC identified in the voxel-based analysis and further restricted within the anatomical EC. These analyses revealed significant periodic modulation only at 6-fold (Figure 2c; t(32) = 3.56, p = 0.006, two-tailed, corrected for multiple comparisons across rotational symmetries; Cohen’s d = 0.62) …”.
We have also revised the “3-fold periodicity in the HPC” subsection of the result as “… ROI analysis, using a functional mask of the HPC identified in the spectral analysis and further restricted within the anatomical HPC, indicated that HPC activity selectively fluctuated at 3-fold periodicity (Figure 3e; t(32) = 3.94, p = 0.002, corrected for multiple comparisons across rotational symmetries; Cohen’s d = 0.70) …”.
(6) Figure 2d: Did you rotationally align 0{degree sign} across participants? Please state explicitly whether (or not) 0{degree sign} aligns with the x-axis in Greeble space.
We thank the reviewer for this helpful question. Yes, before reconstructing the directional tuning curve in Figure 2d, path directions were rotationally aligned for each participant by subtracting the participant-specific grid orientation (ϕ) estimated from the independent dataset (odd sessions). We have now made this description explicit in the revised manuscript in the “6-fold periodicity in the EC” subsection of the Results, stating “… To account for individual difference in spatial phase, path directions were calibrated by subtracting the participant-specific grid orientation estimated from the odd sessions ...”.
(7) Clustering of grid orientations in 30 participants: What does “Bonferroni corrected” refer to? Also, the Rayleigh test is sensitive to the number of voxels - do you obtain the same results when using pair-wise phase consistency?
“Bonferroni corrected” here refers to correction across participants. We have clarified this in the first paragraph of the “6-fold periodicity in the EC” subsection of the Result and in the legend of Supplementary Figure S5 as “Bonferroni-corrected across participants.”
To examine whether our findings were sensitive to the number of voxels, we followed the reviewer’s guidance to compute pairwise phase consistency (PPC; Vinck et al., 2010) for each participant. The PPC results replicated those obtained with the Rayleigh test. We have updated the new results into the Supplementary Figure S5. We also updated the “Statistical Analysis” subsection of the Methods to describe PPC as “For the PPC (Vinck et al., 2010), significance was tested using 5,000 permutations of uniformly distributed random phases (0–2π) to generate a null distribution for comparison with the observed PPC”.
(8) 6-fold periodicity in the EC: Do you compute an average grid orientation across all EC voxels, or do you compute voxel-specific grid orientations?
Following the protocol originally described by Doeller et al. (2010), we estimated voxel-wise grid orientations within the EC and then obtained a participant-specific orientation by averaging across voxels within a hand-drawn bilateral EC mask. The procedure is described in detail in the “Sinusoidal modulation” subsection of the Methods.
(9) Hand-drawn bilateral EC mask: What was your procedure for drawing this mask? What results do you get with a standard mask, for example, from Freesurfer or SPM? Why do you perform this analysis bilaterally, given that the earlier analysis identified 6-fold symmetry only in the right EC? What do you mean by "permutation corrected for multiple comparisons"?
We thank the reviewer for raising these important methodological points. To our knowledge, no standard volumetric atlas provides an anatomically defined entorhinal cortex (EC) mask. For example, the built-in Harvard–Oxford cortical structural atlas in FSL contains only a parahippocampal region that encompasses, but does not isolate, the EC. The AAL atlas likewise does not contain an EC region. In FreeSurfer, an EC label is available, but only in the fsaverage surface space, which is not directly compatible with MNI-based volumetric group-level analyses.
Therefore, we constructed a bilateral EC mask by manually delineating the EC according to the detailed anatomical landmarks described by Insausti et al. (1998). Masks were created using ITK-SNAP (Version 3.8, www.itksnap.org). For transparency and reproducibility, the mask has been made publicly available at the Science Data Bank (link: https://www.scidb.cn/s/NBriAn), as indicated in the revised Data and Code availability section.
Regarding the use of a bilateral EC mask despite voxel-wise effects being strongest in the right EC. First, we did not have any a priori hypothesis regarding laterality of EC involvement before performing analyses. Second, previous studies estimated grid orientation using a bilateral EC mask in their sinusoidal analyses (Doeller et al., 2010; Constantinescu et al., 2016; Bao et al., 2019; Wagner et al., 2023; Raithel et al., 2023). We therefore followed this established approach to estimate grid orientation.
By “permutation corrected for multiple comparisons” we refer to the family-wise error correction applied to the reconstructed directional tuning curves (Figure 2d for the EC, Figure 3f for the HPC). Specifically, directional labels were randomly shuffled 5,000 times, and an FFT was applied to each shuffled dataset to compute spectral power at each fold. This procedure generated null distributions of spectral power for each symmetry. For each fold, the 95th percentile of the maximal power across permutations was used as the uncorrected threshold. To correct across folds, the 95th percentile of the maximal suprathreshold power across all symmetries was taken as the family-wise error–corrected threshold. We have clarified this procedure in the revised “Statistical Analysis” subsection of the Methods.
(10) Figures 3b and 3d: Why do different hippocampal voxels show significance for the sinusoidal versus spectral analysis? Shouldn’t the analyses be redundant and, thus, identify the same significant voxels?
We thank the reviewer for this insightful question. Although both sinusoidal modulation and spectral analysis aim to detect periodic neural activity, the two approaches are methodologically distinct and are therefore not expected to identify exactly the same significant voxels.
Sinusoidal modulation relies on a GLM with sine and cosine regressors to test for phase-aligned periodicity (e.g., 3-fold or 6-fold), calibrated according to the estimated grid orientation. This approach is highly specific but critically depends on accurate orientation estimation. In contrast, spectral analysis applies Fourier decomposition to the directional tuning profile, enabling the detection of periodic components without requiring orientation calibration.
Accordingly, the two analyses are not redundant but complementary. The FFT approach allows for an unbiased exploration of multiple candidate periodicities (e.g., 3–7-fold) without predefined assumptions, thereby providing a critical cross-validation of the sinusoidal GLM results. This strengthens the evidence for 6-fold periodicity in EC and 3-fold periodicity in HPC. Furthermore, FFT uniquely facilitates the analysis of periodicities in behavioral performance data, which is not feasible with standard sinusoidal GLM approaches. This methodological consistency enables direct comparison of periodicities across neural and behavioral domains.
Additionally, the anatomical distributions of the HPC clusters appear more similar between Figure 3b and Figure 3d after re-plotting Figure 3d using the peak voxel coordinates (x = –24, y = –18), which are closer to those used for Figure 3b (x = –24, y = –20), as shown in the revised Figure 3.
Taken together, the two analyses serve distinct but complementary purposes.
(11) 3-fold sinusoidal analysis in hippocampus: What kind of small volume are you using to correct for multiple comparisons?
We thank the reviewer for this comment. The same small volume correction procedure was applied as described in R4. Specifically, the anatomical mask of the bilateral medial temporal lobe (MTL), as defined by the AAL atlas, was used as the small volume for correction. This procedure has been clarified in the revised Statistical Analysis section of the Methods as following: “… with small-volume correction (SVC) applied within the bilateral MTL.”
(12) Figure S5: “right HPC” – isn’t the cluster in the left hippocampus?
We are sorry for the confusion. The brain image was present in radiological orientation (i.e., the left and right orientations are flipped). We also checked the figure and confirmed that the cluster shown in the original Figure S5 (i.e., Figure S6 in the revised manuscript) is correctly labeled as the right hippocampus, as indicated by the MNI coordinate (x = 22), where positive x values denote the right hemisphere. To avoid potential confusion, we have explicitly added the statement “Volumetric results are displayed in radiological orientation” to the figure legends of all volume-based results.
(13) Figure S5: Why are the significant voxels different from the 3-fold symmetry analysis using 10{degree sign} bins?
As shown in R10, the apparent differences largely reflect variation in MNI coordinates. After adjusting for display coordinates, the anatomical locations of the significant clusters are in fact highly similar between the 10°-binned (Figure 3d, shown above) and the 20°-binned results (Figure S6).
Although both analyses rely on sinusoidal modulation, they differ in the resolution of the input angular bins (10° vs. 20°). Combined with the inherent noise in fMRI data, this makes it unlikely that the two approaches would yield exactly the same set of significant voxels. Importantly, both analyses consistently reveal robust 3-fold periodicity in the hippocampus, indicating that the observed effect is not dependent on angular bin size.
(14) Figure 4a and corresponding text: What is the unit? Phase at which frequency? Are you using a circular-circular correlation to test for the relationship?
We thank the reviewer for raising this important point. In the revised manuscript, we have clarified that the unit of the phase values is radians, corresponding to the 6-fold periodic component in the EC and the 3-fold periodic component in the HPC. In the original Figure 4a, both EC and HPC phases—estimated from sinusoidal modulation—were analyzed using Pearson correlation. We have since realized issues with this approach, as also noted R5 to Reviewer #1.
In the revised analysis and Figure 4a (as shown above), we re-evaluated the relationship between EC and HPC phases using a circular–circular correlation (Jammalamadaka & Sengupta, 2001), implemented in the CircStat MATLAB toolbox. The “Phase synchronization between the HPC and EC activity” subsection of the Result has been accordingly updated as following:
“To examine whether the spatial phase structure in one region could predict that in another, we tested whether the orientations of the 6-fold EC and 3-fold HPC periodic activities, estimated from odd-numbered sessions using sinusoidal modulation with rotationally symmetric parameters (in radians), were correlated across participants. A cross-participant circular–circular correlation was conducted between the spatial phases of the two areas to quantify the spatial correspondence of their activity patterns (EC: purple dots; HPC: green dots) (Jammalamadaka & Sengupta, 2001). The analysis revealed a significant circular correlation (Figure 4a; r = 0.42, p < 0.001) …”.
In the “Statistical analysis” subsection of the method:
“… The relationship between EC and HPC phases was evaluated using the circular–circular correlation (Jammalamadaka & Sengupta, 2001) implemented in the CircStat MATLAB toolbox …”.
(15) Paragraph following “We further examined amplitude-phase coupling...” - please clarify what data goes into this analysis.
We thank the reviewer for this helpful comment. In this analysis, the input data consisted of hippocampal (HPC) phase and entorhinal (EC) amplitude, both extracted using the Hilbert transform from the reconstructed BOLD signals of the EC and HPC derived through sinusoidal modulation. We have substantially revised the description of the amplitude–phase coupling analysis in the third paragraph of the “Phase Synchronization Between HPC and EC Activity” subsection of the Results to clarify this procedure.
(16) Alignment between EC 6-fold phases and HC 3-fold phases: Why don't you simply test whether the preferred 6-fold orientations in EC are similar to the preferred 3-fold phases in HC? The phase-amplitude coupling analyses seem sophisticated but are complex, so it is somewhat difficult to judge to what extent they are correct.
We thank the reviewer for this thoughtful comment. We employed two complementary analyses to examine the relationship between EC and HPC activity. In the revised Figure 4 (as shown in Figure 4 for Reviewer #1), Figure 4a provides a direct and intuitive measure of the phase relationship between the two regions using circular–circular correlation. Figure 4b–c examines whether the activity peaks of the two regions are aligned across path directions using cross-frequency amplitude–phase coupling, given our hypothesis that the spatial phase of the HPC depends on EC projections. These two analyses are complementary: a phase correlation does not necessarily imply peak-to-peak alignment, and conversely, peak alignment does not always yield a statistically significant phase correlation. We therefore combined multiple analytical approaches as a cross-validation across methods, providing convergent evidence for robust EC–HPC coupling.
(17) Figure 5: Do these results hold when you estimate performance just based on “deviation from the goal to ending locations” (without taking path length into account)?
We thank the reviewer for this thoughtful suggestion. Following the reviewer’s advice, we re-estimated behavioral performance using the deviation between the goal and ending locations (i.e., error size) and path length independently. As shown in the new Figure S9, no significant periodicity was observed in error size (p > 0.05), whereas a robust 3-fold periodicity was found for path length (p < 0.05, corrected for multiple comparisons).
We employed two behavioral metrics,(1) path length and (2) error size, for complementary reasons. In our task, participants navigated using four discrete keys corresponding to the cardinal directions (north, south, east, and west). This design inherently induces a 4-fold bias in path directions, as described in the “Behavioral performance” subsection of the Methods. To minimize this artifact, we computed the objectively optimal path length and used it to calibrate participants’ path lengths. However, error size could not be corrected in the same manner and retained a residual 4-fold tendency (see Figure S9d).
Given that both path length and error size are behaviorally relevant and capture distinct aspects of task performance, we decided to retain both measures when quantifying behavioral periodicity. This clarification has been incorporated into the “Behavioral performance” subsection of the Methods, and the 2<sup>nd</sup> paragraph of the “3-fold periodicity in human behavior” subsection of the Results.
(18) Phase locking between behavioral performance and hippocampal activity: What is your way of creating surrogates here?
We thank the reviewer for this helpful question. Surrogate datasets were generated by circularly shifting the signal series along the direction axis across all possible offsets (following Canolty et al., 2006). This procedure preserves the internal phase structure within each domain while disrupting consistent phase alignment, thereby removing any systematic coupling between the two signals. Each surrogate dataset underwent identical filtering and coherence computation to generate a null distribution, and the observed coherence strength was compared with this distribution using paired t-tests across participants. The statistical analysis section has been systematically revised to incorporate these methodological details.
(19) I could not follow why the authors equate 3-fold symmetry with vectorial representations. This includes statements such as “these empirical findings provide a potential explanation for the formation of vectorial representation observed in the HPC.” Please clarify.
We thank the reviewer for raising this point. Please refer to our response to R2 for Reviewer #1 and the revised Introduction (paragraphs 2–4), where we explicitly explain why the three primary axes of the hexagonal grid cell code can manifest as vector fields.
(20) It was unclear whether the sentence “The EC provides a foundation for the formation of periodic representations in the HPC” is based on the authors’ observations or on other findings. If based on the authors’ findings, this statement seems too strong, given that no other studies have reported periodic representations in the hippocampus to date (to the best of my knowledge).
We thank the reviewer for this comment. We agree that the original wording lacked sufficient rigor. We have extensively revised the 3rd paragraph of the Discussion section with more cautious language by reducing overinterpretation and emphasizing the consistency of our findings with prior empirical evidence, as follows: “The EC–HPC PhaseSync model demonstrates how a vectorial representation may emerge in the HPC from the projections of populations of periodic grid codes in the EC. The model was motivated by two observations. First, the EC intrinsically serves as the major source of hippocampal input (Witter and Amaral, 1991; van Groen et al., 2003; Garcia and Buffalo, 2020), and grid codes exhibit nearly invariant spatial orientations (Hafting et al., 2005; Gardner et al., 2022). Second, mental planning, characterized by “forward replay” (Dragoi and Tonegawa, 2011; Pfeiffer, 2020), has the capacity to activate populations of grid cells that represent sequential experiences in the absence of actual physical movement (Nyberg et al., 2022). We hypothesize that an integrated path code of sequential experiences may eventually be generated in the HPC, providing a vectorial gradient toward the goal location. The path code exhibits regular, vector-like representations when the path direction aligns with the orientations of grid axes, and becomes irregular when they misalign. This explanation is consistent with the band-like representations observed in the dorsomedial EC (Krupic et al., 2012) and the irregular activity fields of trace cells in the HPC (Poulter et al., 2021). ”
-
-
-
Reading back all those old posts and weeknotes I have here is super nice and reminds me:Keeping a record of things is really valuable. Just write and trust that it will come in handy at some point.I used to do so many things in a given week. Compared to what I’m doing now, my life was insanely eventful.I was consistently (too) early on a lot of things. For instance: I read myself complaining about restaurants and food in Amsterdam, something which is mostly solved now.
Like myself Alper is his own most frequent reader of his blog. Mentions realising how much he did in a week earlier in his life. Same for me, things that now might be a big thing in a week, were Tuesday afternoon 15yrs ago. It's not just age I suspect, but also an overall attenuation that Covid brought?
Tags
Annotators
URL
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the original reviews
A point by point response included below. Before we turn to that we want to note one change that we decided to introduce, related to generalization on unseen tissues/cell types (Figure 3a in the original submission and related question by Reviewer #2 below). This analysis was based on adding a latent “RBP state” representation during learning of condition/tissue specific splicing. The “RBP state” per condition is captured by a dedicated encoder. Our original plan was to have a paper describing a new RBP-AE model we developed in parallel, which also served as the base to capture this “RBP State”. However, we got delayed in getting this second paper finalized (it was led by other lab members, some of whom have already left the lab). This delay affected the TrASPr manuscript as TrASPr’s code should be available and analysis reproducible upon publication. After much deliberation, we decided that in order to comply with reproducibility standards while not self scooping the RBP-AE paper, we eventually decided to take out the RBP-AE and replace it with a vanilla PCA based embedding for the “RBP-State”. The PCA approach is simpler and reproducible, based on linear transformation of the RBPs expression vector into a lower dimension. The qualitative results included in Figure 3a still hold, and we also produced the new results suggested by Reviewer #2 in other GTEX tissues with this PCA based embedding (below).
We don’t believe the switch to PCA based embedding should have any bearing on the current manuscript evaluation but wanted to take this opportunity to explain the reasoning behind this additional change.
Public Reviews:
Reviewer #1 (Public review):
Summary:
The authors propose a transformer-based model for the prediction of condition - or tissue-specific alternative splicing and demonstrate its utility in the design of RNAs with desired splicing outcomes, which is a novel application. The model is compared to relevant existing approaches (Pangolin and SpliceAI) and the authors clearly demonstrate its advantage. Overall, a compelling method that is well thought out and evaluated.
Strengths:
(1) The model is well thought out: rather than modeling a cassette exon using a single generic deep learning model as has been done e.g. in SpliceAI and related work, the authors propose a modular architecture that focuses on different regions around a potential exon skipping event, which enables the model to learn representations that are specific to those regions. Because each component in the model focuses on a fixed length short sequence segment, the model can learn position-specific features. Another difference compared to Pangolin and SpliceAI which are focused on modeling individual splice junctions is the focus on modeling a complete alternative splicing event.
(2) The model is evaluated in a rigorous way - it is compared to the most relevant state-of-the-art models, uses machine learning best practices, and an ablation study demonstrates the contribution of each component of the architecture.
(3) Experimental work supports the computational predictions.
(4) The authors use their model for sequence design to optimize splicing outcomes, which is a novel application.
We wholeheartedly thank Reviewer #1 for these positive comments regarding the modeling approach we took to this task and the evaluations we performed. We have put a lot of work and thought into this and it is gratifying to see the results of that work acknowledged like this.
Weaknesses:
No weaknesses were identified by this reviewer, but I have the following comments:
(1) I would be curious to see evidence that the model is learning position-specific representations.
This is an excellent suggestion to further assess what the model is learning. To get a better sense of the position-specific representation we performed the following analyses:
(1) Switching the transformers relative order: All transformers are pretrained on 3’ and 5’ splice site regions before fine-tunning for the PSI and dPSI prediction task. We hypothesized that if relative position is important, switching the order of the transformers would make a large difference on prediction accuracy. Indeed if we switch the 3’ and 5’ we see as expected a severe drop in performance, with Pearson correlation on test data dropping from 0.82 to 0.11. Next, we switched the two 5’ and 3’ transformers, observing a drop to 0.65 and 0.78 respectively. When focusing only on changing events the drop was from 0.66 to 0.54 (for 3’ SS transformers), 0.48 (for 5’ SS transformers), and 0.13 (when the 3’ and 5’ transformers flanking the alternative exon were switched).
(2) Position specific effect of RBPs: We wanted to test whether the model is able to learn position specific effects for RBPs. For this we focused on two RBPs, FOX (a family of three highly related RBPs), and QKI, both have a relatively well defined motif, known condition and position specific effect identified via RBP KD experiments combined with CLIP experiments (e.g. PMID: 23525800, PMID: 24637117, PMID: 32728246). For each, we randomly selected 40 highly and 40 lowly included cassette exons sequences. We then ran in-silico mutagenesis experiments where we replaced small windows of sequences with the RBP motifs (80 for RBFOX and 80 for QKI), then compared TrASPR’s predictions for the average predictions for 5 random sequences inserted in the same location. The results of this are now shown in Figure 4 Supp 3, where the y-axis represents the dPSI effect per position (x-axis), and the color represents the percentile of observed effects over inserting motifs in that position across all 80 sequences tested. We see that both RBPs have strong positional preferences for exerting a strong effect on the alternative exon. We also see differences between binding upstream and downstream of the alternative exon. These results, learned by the model from natural tissue-specific variations, recapitulate nicely the results derived from high-throughput experimental assays. However, we also note that effects were highly sequence specific. For example, RBFOX is generally expected to increase inclusion when binding downstream of the alternative exon and decrease inclusion when binding upstream. While we do observe such a trend we also see cases where the opposite effects are observed. These sequence specific effects have been reported in the literature but may also represent cases where the model errs in the effect’s direction. We discuss these new results in the revised text.
(3) Assessing BOS sequence edits to achieve tissue-specific splicing: Here we decided to test whether BOS edits in intronic regions (at least 8b away from the nearest splice site) are important for the tissue-specific effect. The results are now included in Figure 6 Supp 1, clearly demonstrating that most of the neuronal specific changes achieved by BOS were based on changing the introns, with a strong effect observed for both up and downstream intron edits.
(2) The transformer encoders in TrASPr model sequences with a rather limited sequence size of 200 bp; therefore, for long introns, the model will not have good coverage of the intronic sequence. This is not expected to be an issue for exons.
The reviewer is raising a good question here. On one hand, one may hypothesize that, as the reviewer seems to suggest, TrASPr may not do well on long introns as it lacks the full intronic sequence.
Conversely, one may also hypothesize that for long introns, where the flanking exons are outside the window of SpliceAI/Pangolin, TrASPr may have an advantage.
Given this good question and a related one by Reviewer #2, we divided prediction accuracy by intron length and the alternative exon length.
For short exons (<100bp) we find TrASPr and Pangolin perform similarly, but for longer exons, especially those > 200, TrASPr results are better. When dividing samples by the total length of the upstream and downstream intron, we find TrASPr outperform all other models for introns of combined length up to 6K, but Pangolin gets better results when the combined intron length is over 10K. This latter result is interesting as it means that contrary to the second hypothesis laid out above, Pangolin’s performance did not degrade for events where the flanking exons were outside its field of view. We note that all of the above holds whether we assess all events or just cases of tissue specific changes. It is interesting to think about the mechanistic causes for this. For example, it is possible that cassette exons involving very long introns evoke a different splicing mechanism where the flanking exons are not as critical and/or there is more signal in the introns which is missed by TrASPr. We include these new results now as Figure 2 - Supp 1,2 and discuss these in the main text.
(3) In the context of sequence design, creating a desired tissue- or condition-specific effect would likely require disrupting or creating motifs for splicing regulatory proteins. In your experiments for neuronal-specific Daam1 exon 16, have you seen evidence for that? Most of the edits are close to splice junctions, but a few are further away.
That is another good question. Regarding Daam1 exon 16, in the original paper describing the mutation locations some motif similarities were noted to PTB (CU) and CUG/Mbnl-like elements (Barash et al Nature 2010). In order to explore this question beyond this specific case we assessed the importance of intronic edits by BOS to achieve a tissue specific splicing profile - see above.
(4) For sequence design, of tissue- or condition-specific effect in neuronal-specific Daam1 exon 16 the upstream exonic splice junction had the most sequence edits. Is that a general observation? How about the relative importance of the four transformer regions in TrASPr prediction performance?
This is another excellent question. Please see new experiments described above for RBP positional effect and BOS edits in intronic regions which attempt to give at least partial answers to these questions. We believe a much more systematic analysis can be done to explore these questions but such evaluation is beyond the scope of this work.
(5) The idea of lightweight transformer models is compelling, and is widely applicable. It has been used elsewhere. One paper that came to mind in the protein realm:
Singh, Rohit, et al. "Learning the language of antibody hypervariability." Proceedings of the National Academy of Sciences 122.1 (2025): e2418918121.
We definitely do not make any claim this approach of using lighter, dedicated models instead of a large ‘foundation’ model has not been taken before. We believe Rohit et al mentioned above represents a somewhat different approach, where their model (AbMAP) fine-tunes large general protein foundational models (PLM) for antibody-sequence inputs by supervising on antibody structure and binding specificity examples. We added a description of this modeling approach citing the above work and another one which specifically handles RNA splicing (intron retention, PMID: 39792954).
Reviewer #2 (Public review):
Summary:
The authors present a transformer-based model, TrASPr, for the task of tissue-specific splicing prediction (with experiments primarily focused on the case of cassette exon inclusion) as well as an optimization framework (BOS) for the task of designing RNA sequences for desired splicing outcomes.
For the first task, the main methodological contribution is to train four transformer-based models on the 400bp regions surrounding each splice site, the rationale being that this is where most splicing regulatory information is. In contrast, previous work trained one model on a long genomic region. This new design should help the model capture more easily interactions between splice sites. It should also help in cases of very long introns, which are relatively common in the human genome.
TrASPr's performance is evaluated in comparison to previous models (SpliceAI, Pangolin, and SpliceTransformer) on numerous tasks including splicing predictions on GTEx tissues, ENCODE cell lines, RBP KD data, and mutagenesis data. The scope of these evaluations is ambitious; however, significant details on most of the analyses are missing, making it difficult to evaluate the strength of the evidence. Additionally, state-of-the-art models (SpliceAI and Pangolin) are reported to perform extremely poorly in some tasks, which is surprising in light of previous reports of their overall good prediction accuracy; the reasoning for this lack of performance compared to TrASPr is not explored.
In the second task, the authors combine Latent Space Bayesian Optimization (LSBO) with a Transformer-based variational autoencoder to optimize RNA sequences for a given splicing-related objective function. This method (BOS) appears to be a novel application of LSBO, with promising results on several computational evaluations and the potential to be impactful on sequence design for both splicing-related objectives and other tasks.
We thank Reviewer #2 for this detailed summary and positive view of our work. It seems the main issue raised in this summary regards the evaluations: The reviewer finds details of the evaluations missing and the fact that SpliceAI and Pangolin perform poorly on some of the tasks to be surprising. We made a concise effort to include the required details, including code and data tables. In short, some of the concerns were addressed by adding additional evaluations, some by clarifying missing details, and some by better explaining where Pangolin and SpliceAI may excel vs. settings where these may not do as well. More details are given below.
Strengths:
(1) A novel machine learning model for an important problem in RNA biology with excellent prediction accuracy.
(2) Instead of being based on a generic design as in previous work, the proposed model incorporates biological domain knowledge (that regulatory information is concentrated around splice sites). This way of using inductive bias can be important to future work on other sequence-based prediction tasks.
Weaknesses:
(1) Most of the analyses presented in the manuscript are described in broad strokes and are often confusing. As a result, it is difficult to assess the significance of the contribution.
We made an effort to make the tasks be specific and detailed, including making the code and data of those available. We believe this helped improve clarity in the revised version.
(2) As more and more models are being proposed for splicing prediction (SpliceAI, Pangolin, SpliceTransformer, TrASPr), there is a need for establishing standard benchmarks, similar to those in computer vision (ImageNet). Without such benchmarks, it is exceedingly difficult to compare models. For instance, Pangolin was apparently trained on a different dataset (Cardoso-Moreira et al. 2019), and using a different processing pipeline (based on SpliSER) than the ones used in this submission. As a result, the inferior performance of Pangolin reported here could potentially be due to subtle distribution shifts. The authors should add a discussion of the differences in the training set, and whether they affect your comparisons (e.g., in Figure 2). They should also consider adding a table summarizing the various datasets used in their previous work for training and testing. Publishing their training and testing datasets in an easy-to-use format would be a fantastic contribution to the community, establishing a common benchmark to be used by others.
There are several good points to unpack here. Starting from the last one, we very much agree that a standard benchmark will be useful to include. For tissue specific splicing quantification we used the GTEx dataset from which we select six representative human tissues (heart, cerebellum, lung, liver, spleen, and EBV-transformed lymphocytes). In total, we collected 38394 cassette exon events quantified across 15 samples (here a ‘sample’ is a cassette exon quantified in two tissues) from the GTEx dataset with high-confidence quantification for their PSIs based on MAJIQ. A detailed description of how this data was derived is now included in the Methods section, and the data itself is made available via the bitbucket repository with the code.
Next, regarding the usage of different data and distribution shifts for Pangolin: The reviewer is right to note there are many differences between how Pangolin and TrASPr were trained. This makes it hard to determine whether the improvements we saw are not just a result of different training data/labels. To address this issue, we first tried to finetune the pre-trained Pangolin with MAJIQ’s PSI dataset: we use the subset of the GTEx dataset described above, focusing on the three tissues analyzed in Pangolin’s paper—heart, cerebellum, and liver—for a fair comparison. In total, we obtained 17,218 events, and we followed the same training and test split as reported in the Pangolin paper. We got Pearson: 0.78 Spearman: 0.68 which are values similar to what we got without this extra fine tuning. Next, we retrained Pangolin from scratch, with the full tissues and training set used for TrASPr, which was derived from MAJIQ’s quantifications. Since our model only trained on human data with 6 tissues at the same time, we modified Pangolin from original 4 splice site usage outputs to 6 PSI outputs. We tried to take the sequence centered with the first or the second splice site of the mid exon. This test resulted in low performance (3’ SS: pearson 0.21 5’ SS: 0.26.).
The above tests are obviously not exhaustive but their results suggest that the differences we observe are unlikely to be driven by distribution shifts. Notably, the original Pangolin was trained on much more data (four species, four tissues each, and sliding windows across the entire genome). This training seems to be important for performance while the fact we switched from Pangolin’s splice site usage to MAJIQ’s PSI was not a major contributor. Other potential reasons for the improvements we observed include the architecture, target function, and side information (see below) but a complete delineation of those is beyond the scope of this work.
(3) Related to the previous point, as discussed in the manuscript, SpliceAI, and Pangolin are not designed to predict PSI of cassette exons. Instead, they assign a "splice site probability" to each nucleotide. Converting this to a PSI prediction is not obvious, and the method chosen by the authors (averaging the two probabilities (?)) is likely not optimal. It would be interesting to see what happens if an MLP is used on top of the four predictions (or the outputs of the top layers) from SpliceAI/Pangolin. This could also indicate where the improvement in TrASPr comes from: is it because TrASPr combines information from all four splice sites? Also, consider fine-tuning Pangolin on cassette exons only (as you do for your model).
Please see the above response. We did not investigate more sophisticated models that adjust Pangolin’s architecture further as such modifications constitute new models which are beyond the scope of this work.
(4) L141, "TrASPr can handle cassette exons spanning a wide range of window sizes from 181 to 329,227 bases - thanks to its multi-transformer architecture." This is reported to be one of the primary advantages compared to existing models. Additional analysis should be included on how TrASPr performs across varying exon and intron sizes, with comparison to SpliceAI, etc.
This was a good suggestion, related to another comment made by Reviewer #1. Please see above our response to them with a breakdown by exon/intron length.
(5) L171, "training it on cassette exons". This seems like an important point: previous models were trained mostly on constitutive exons, whereas here the model is trained specifically on cassette exons. This should be discussed in more detail.
Previous models were not trained exclusively on constitutive exons and Pangolin specifically was trained with their version of junction usage across tissues. That said, the reviewer’s point is valid (and similar to ones made above) about a need to have a matched training/testing and potential distribution shifts. Please see response and evaluations described above.
(6) L214, ablations of individual features are missing.
These were now added to the table which we moved to the main text (see table also below).
(7) L230, "ENCODE cell lines", it is not clear why other tissues from GTEx were not included.
Good question. The task here was to assess predictions in unseen conditions, hence we opted to test on completely different data of human cell lines rather than additional tissue samples. Following the reviewers suggestion we also evaluated predictions on two additional GTEx tissues, Cortex and Adrenal Gland. These new results, as well as the previous ones for ENCODE, were updated to use the PCA based embedding of “RBP-State” as described above. We also compared the predictions using the PCA based embedding of the “RBP-State” to training directly on data (not the test data of course) from these tissues. See updated Figure 3a,b. Figure 3 Supp 1,2.
(8) L239, it is surprising that SpliceAI performs so badly, and might suggest a mistake in the analysis. Additional analysis and possible explanations should be provided to support these claims. Similarly, the complete failure of SpliceAI and Pangolin is shown in Figure 4d.
Line 239 refers to predicting relative inclusion levels between competing 3’ and 5’ splice sites. We admit we too expected this to be better for SpliceAI and Pangolin but we were not able to find bugs in our analysis (which is all made available for readers and reviewers alike). Regarding this expectation to perform better, first we note that we are not aware of a similar assessment being done for either of those algorithms (i.e. relative inclusion for 3’ and 5’ alternative splice site events). Instead, our initial expectation, and likely the reviewer’s as well, was based on their detection of splice site strengthening/weakening due to mutations, including cryptic splice site activation. More generally though, it is worth noting in this context that given how SpliceAI, Pangolin and other algorithms have been presented in papers/media/scientific discussions, we believe there is a potential misperception regarding tasks that SpliceAI and Pangolin excel at vs other tasks where they should not necessarily be expected to excel. Both algorithms focus on cryptic splice site creation/disruption. This has been the focus of those papers and subsequent applications. While Pangolin added tissue specificity to SpliceAI training, the authors themselves admit “...predicting differential splicing across tissues from sequence alone is possible but remains a considerable challenge and requires further investigation”. The actual performance on this task is not included in Pangolin’s main text, but we refer Reviewer #2 to supplementary figure S4 in the Pangolin manuscript to get a sense of Pangolin’s reported performance on this task. Similar to that, Figure 4d in our manuscript is for predicting ‘tissue specific’ regulators. We do not think it is surprising that SpliceAI (tissue agnostic) and Pangolin (slight improvement compared to SpliceAI in tissue specific predictions) do not perform well on this task. Similarly, we do not find the results in Figure 4C surprising either. These are for mutations that slightly alter inclusion level of an exon, not something SpliceAI was trained on - SpiceAI was trained on genomic splice sites with yes/no labels across the genome. As noted elsewhere in our response, re-training Pangolin on this mutagenesis dataset results in performance much closer to that of TrASPr. That is to be expected as well - Pangolin is constructed to capture changes in PSI (or splice site usage as defined by the authors), those changes are not even tissue specific for the CD19 data and the model has no problem/lack of capacity to generalize from the training set just like TrASPr does. In fact, if you only use combinations of known mutations seen during training a simple regression model gives correlation of ~92-95% (Cortés-López et al 2022). In summary, we believe that better understanding of what one can realistically expect from models such as SpliceAI, Pangolin, and TrASPr will go a long way to have them better understood and used effectively. We have tried to make this more clear in the revision.
(9) BOS seems like a separate contribution that belongs in a separate publication. Instead, consider providing more details on TrASPr.
We thank the reviewer for the suggestion. We agree those are two distinct contributions/algorithms and we indeed considered having them as two separate papers. However, there is strong coupling between the design algorithm (BOS) and the predictor that enables it (TrASPr). This coupling is both conceptual (TrASPr as a “teacher”) and practical in terms of evaluations. While we use experimental data (experiments done involving Daam1 exon 16, CD19 exon 2) we still rely heavily on evaluations by TrASPr itself. A completely independent evaluation would have required a high-throughput experimental system to assess designs, which is beyond the scope of the current paper. For those reasons we eventually decided to make it into what we hope is a more compelling combined story about generative models for prediction and design of RNA splicing.
(10) The authors should consider evaluating BOS using Pangolin or SpliceTransformer as the oracle, in order to measure the contribution to the sequence generation task provided by BOS vs TrASPr.
We can definitely see the logic behind trying BOS with different predictors. That said, as we note above most of BOS evaluations are based on the “teacher”. As such, it is unclear what value replacing the teacher would bring. We also note that given this limitation we focus mostly on evaluations in comparison to existing approaches (genetic algorithm or random mutations as a strawman).
Recommendations for the authors:
Reviewer #1 (Recommendations for the authors):
Additional comments:
(1) Is your model picking up transcription factor binding sites in addition to RBPs? TFs have been recently shown to have a role in splicing regulation:
Daoud, Ahmed, and Asa Ben-Hur. "The role of chromatin state in intron retention: A case study in leveraging large scale deep learning models." PLOS Computational Biology 21.1 (2025): e1012755.
We agree this is an interesting point to explore, especially given the series of works from the Ben-Hur’s group. We note though that these works focus on intron retention (IR) which we haven’t focused on here, and we only cover short intronic regions flanking the exons. We leave this as a future direction as we believe the scope of this paper is already quite extensive.
(2) SpliceNouveau is a recently published algorithm for the splicing design problem:
Wilkins, Oscar G., et al. "Creation of de novo cryptic splicing for ALS and FTD precision medicine." Science 386.6717 (2024): 61-69.
Thank you for pointing out Wilkins et al recent publication, we now refer to it as well.
(3) Please discuss the relationship between your model and this deep learning model. You will also need to change the following sentence: "Since the splicing sequence design task is novel, there are no prior implementations to reference."
We revised this statement and now refer to several recent publications that propose similar design tasks.
(4) I would suggest adding a histogram of PSI values - they appear to be mostly close to 1 or 0.
PSI values are indeed typically close to either 0 or 1. This is a known phenomenon illustrated in previous studies of splicing (e.g. Shen et al NAR 2012 ). We are not sure what is meant by the comment to add a histogram but we made sure to point this out in the main text:
“...Still, those statistics are dominated by extreme values, such that 33.2\% are smaller than 0.15 and 56.0\% are higher than 0.85. Furthermore, most cassette exons do not change between a given tissue pair (only 14.0\% of the samples in the dataset, \ie a cassette exon measured across two tissues, exhibit ΔΨ| ≥ 0.15).”
(5) Part of the improvement of TrASPr over Pangolin could be the result of a more extensive dataset.
Please see above responses and new analysis.
(6) In the discussion of the roles of alternative splicing, protein diversity is mentioned, but I suggest you also mention the importance of alternative splicing as a regulatory mechanism:
Lewis, Benjamin P., Richard E. Green, and Steven E. Brenner. "Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans." Proceedings of the National Academy of Sciences 100.1 (2003): 189-192.
Thank you for the suggestion. We added that point and citation.
(7) Line 96: You use dPSI without defining it (although quite clear that it should be Delta PSI).
Fixed.
(8) Pretrained transformers: Have you trained separate transformers on acceptor and donor sites, or a single splice junction transformer?
Single splice junction pre-training.
(9) "TrASPr measures the probability that the splice site in the center of Se is included in some tissue" - that's not my understanding of what TrASPr is designed to do.
We revised the above sentence to make it more precise: “Given a genomic sequence context S<sub>e</sub> = (s<sub>e</sub>,...,s<sub>e</sub>), made of a cassette exon e and flanking intronic/exonic regions, TrASPr predicts for tissue c the fraction of transcripts where exon e is included or skipped over, ΔΨ-<sub>e,c,c’</sub>.”
(10) Please include the version of the human genome annotations that you used.
We used GENCODE v40 human genome hg38- this is now included in the Data section.
(11) I did not see a description of the RBP-AE component in the methods section. A bit more detail on the model would be useful as well.
Please see above details about replacing RBP-AE with a simpler linear PCA “RBP-State” encoding. We added details about how the PCA was performed to the Methods section.
(12) Typos, grammar:
- Fix the following sentence: ATP13A2, a lysosomal transmembrane cation transporter, linked to an early-onset form of Parkinson's Disease (PD) when 306 loss-of-function mutations disrupt its function.
Sentence was fixed to now read: “The first example is of a brain cerebellum-specific cassette exon skipping event predicted by TrASPr in the ATP13A2 gene (aka PARK9). ATP13A2 is a lysosomal transmembrane cation transporter, for which loss of function mutation has been linked to early-onset of Parkinson’s Disease (PD)”.
- Line 501: "was set to 4e−4"(the - is a superscript).
Fixed
- A couple of citations are missing in lines 580 and 581.
Thank you for catching this error. Citations in line 580, 581 were fixed.
(13) Paper title: Generative modeling for RNA splicing predictions and design - it would read better as "Generative modeling for RNA splicing prediction and design", as you are solving the problems of splicing prediction and splicing design.
Thank you for the suggestion. We updated the title and removed the plural form.
Reviewer #2 (Recommendations for the authors):
(1) Appendices are not very common in biology journals. It is also not clear what purpose the appendix serves exactly - it seems to repeat some of the things said earlier. Consider merging it into the methods or the main text.
We merged the appendices into the Methods section and removed redundancy.
(2) L112, "For instance, the model could be tasked with designing a new version of the cassette exon, restricted to no more than N edit locations and M total base changes." How are N and M different? Is there a difference between an edit location and a base change?
Yes, N is the number of locations (one can think of it as a start position) of various lengths (e.g. a SNP is of length 1) and the total number of positions edited is M. The text now reads “For instance, the model could be tasked with designing a new version of the cassette exon, restricted to no more than $N$ edit locations (\ie start position of one or more consecutive bases) and $M$ total base changes.”
(3) L122: "DEN was developed for a distinct problem". What prevents one from adapting DEN to your sequence design task? The method should be generic. I do not see what "differs substantially" means here. (Finally, wasn't DEN developed for the task you later refer to as "alternative splice site" (as opposed to "splice site selection")? Use consistent terminology. And in L236 you use "splice site variation" - is that also the same?).
Indeed, our original description was not clear/precise enough. DEN was designed and trained for two tasks: APA, and 5’ alternative splice site usage. The terms “selection”, “usage”, and “variation” were indeed used interchangeably in different locations and the reviewer was right, noting the lack of precision. We have now revised the text to make sure the term “relative usage” is used.
Nonetheless, we hold DEN was indeed defined for different tasks. See figures from Figure 2A, 6A of Linder et al 2020 (the reference was also incorrect as we cited the preprint and not the final paper):
In both cases DEN is trying to optimize a short region for selecting an alternative PA site (left) or a 5’ splice site (right). This work focused on an MPRA dataset of short synthetic sequences inserted in the designated region for train/test. We hold this is indeed a different type of data and task then the one we focus on here. Yes, one can potentially adopt DEN for our task, but this is beyond the scope of this paper. Finally, we note that a more closely related algorithm recently proposed is Ledidi (Schreiber et al 2025) which was posted as a pre-print. Similar to BOS, Ledidi tries to optimize a given sequence and adopt it with a few edits for a given task. Regardless, we updated the main text to make the differences between DEN and the task we defined here for BOS more clear, and we also added a reference to Ledidi and other recent works in the discussion section.
(4) L203, exons with DeltaPSI very close to 0.15 are going to be nearly impossible to classify (or even impossible, considering that the DeltaPSI measurements are not perfect). Consider removing such exons to make the task more feasible.
Yes, this is how it was done. As described in more details below, we defined changing samples as ones where the change was >= 0.15 and non-changing as ones where the change in PSI was < 0.05 to avoid ambiguous cases affecting the classification task.
(5) L230, RBP-AE is not explained in sufficient detail (and does not appear in the methods, apparently). It is not clear how exactly it is trained on each new cellular condition.
Please see response in the opening of this document and Q11 from
Reviewer 1
(6) L230, "significantly improving": the r value actually got worse; it is therefore not clear you can claim any significant improvement. Please mention that fact in the text.
This is a fair point. We note that we view the “a” statistic as potentially more interesting/relevant here as the Pearson “r” is dominated by points being generally close to 0/1. Regardless, revisiting this we realized one can also make a point that the term “significant” is imprecise/misplaced since there is no statistical test done here (side note: given the amount of points, a simple null of same distribution yes/no would pass significance but we don’t think this is an interesting/relevant test here). Also, we note that with the transition to PCA instead of RBP-AE we actually get improvements in both a and r values, both for the ENCODE samples shown in Figure 3a and the two new GTEX tissues we tested (see above). We now changed the text to simply state:
“...As shown in Figure 3a, this latent space representation allows TrSAPr to generalize from the six GTEX tissues to unseen conditions, including unseen GTEX tissues (top row), and ENCODE cell lines (bottom row). It improves prediction accuracy compared to TrASPr lacking PCA (eg a=88.5% vs a=82.3% for ENCODE cell lines), though naturally training on the additional GTEX and ENCODE conditions can lead to better performance (eg a=91.7%, for ENCODE, Figure 3a left column).”
(7) L233, "Notably, previous splicing codes focused solely on cassette exons", Rosenberg et al. focused solely on alternative splice site choice.
Right - we removed that sentence..
(8) L236, "trained TrASPr on datasets for 3' and 5' splice site variations". Please provide more details on this task. What is the input to TrASPr and what is the prediction target (splice site usage, PSI of alternative isoforms)? What datasets are used for this task?
The data for this data was the same GTEx tissue data processed, just for alternative 3’ and 5’ splice sites events. We revised the description of this task in the main task and added information in the Methods section. The data is also included in the repo.
(9) L243, "directly from genomic sequences", and conservation?
Yes, we changed the sentence to read “...directly from genomic sequences combined with related features”
(10) L262, what is the threshold for significant splicing changes?
The threshold is 0.15 We updated the main text to read the following:
The total number of mutations hitting each of the 1198 genomic positions across the 6106 sequences is shown in \FIG{mut_effect}b (left), while the distribution of effects ($|\Delta \Psi|$) observed across those 6106 samples is shown in \FIG{mut_effect}b (right). To this data we applied three testing schemes. The first is a standard 5-fold CV where 20\% of combinations of point mutations were hidden in every fold while the second test involved 'unseen mutation' (UM) where we hide any sample that includes mutations in specific positions for a total of 1480 test samples. As illustrated by the CDF in \FIG{mut_effect}b, most samples (each sample may involve multiple positions mutated) do not involve significant splicing changes. Thus, we also performed a third test using only the 883 samples were mutations cause significant changes ($|\Delta \Psi|\geq 0.15 $).
(11) L266, Pangolin performance is only provided for one of the settings (and it is not clear which). Please provide details of its performance in all settings.
The description was indeed not clear. Pangolin’s performance was similar to SpliceAI as mentioned above but retraining it on the CD19 data yielded much closer performance to TrASPr. We include all the matching tests for Pangolin after retraining in Figure 4 Supp Figure 1.
(12) Please specify "n=" in all relevant plots.
Fixed.
(13) Figure 3a, "The tissues were first represented as tokens, and new cell line results were predicted based on the average over conditions during training." Please explain this procedure in more detail. What are these tokens and how are they provided to the model? Are the cell line predictions the average of the predictions for the training tissues?
Yes, we compared to simply the average over the predictions for the training tissues for that specific event as baseline to assess improvements (see related work pointing for the need to have similar baselines in DL for genomics in https://pubmed.ncbi.nlm.nih.gov/33213499/). Regarding the tokens - we encode each tissue type as a possible value and feed the two tissues as two tokens to the transformer.
(14) Figure 4b, the total count in the histogram is much greater than 6106. Please explain the dataset you're using in more detail, and what exactly is shown here.
We updated the text to read:
“...we used 6106 sequence samples where each sample may have multiple positions mutated (\ie mutation combinations) in exon 2 of CD19 and its flanking introns and exons (Cortes et al 2022). The total number of mutations hitting each of the 1198 genomic positions across the 6106 sequences is shown in Figure 4b (left).”
(15) Figure 5a, how are the prediction thresholds (TrASPr passed, TrASPr stringent, and TrASPr very stringent) defined?
Passed: dpsi>0.1, Stringent: dpsi>0.15, Very stringent: dpsi>0.2 This is now included in the main text.
(16) L417, please include more detail on the relative size of TrASPr compared to other models (e.g. number of parameters, required compute, etc.).
SpliceAI is a general-purpose splicing predictor with 32-layer deep residual neural network to capture long-range dependencies in genomic sequences. Pangolin is a deep learning model specifically designed for predicting tissue-specific splicing with similar architecture as SpliceAI. The implementation of SpliceAI that can be found here https://huggingface.co/multimolecule/spliceai involves an ensemble of 5 such models for a total of ~3.5M parameters. TrASPr, has 4 BERT transformers (each 6 layers and 12 heads) and MLP a top of those for a total of ~189M parameters. Evo 2, a genomic ‘foundation’ model has 40B parameters, DNABERT has ~86M (a single BERT with 12 layers and 12 heads), and Borzoi has 186M parameters (as stated in https://www.biorxiv.org/content/10.1101/2025.05.26.656171v2). We note that the difference here is not just in model size but also the amount of data used to train the model. We edited the original L417 to reflect that.
(17) L546, please provide more detail on the VAE. What is the dimension of the latent representation?
We added more details in the Methods section like the missing dimension (256) and definitions for P(Z) and P(S).
(18) Consider citing (and possibly comparing BOS to) Ghari et al., NeurIPS 2024 ("GFlowNet Assisted Biological Sequence Editing").
Added.
(19) Appendix Figure 2, and corresponding main text: it is not clear what is shown here. What is dPSI+ and dPSI-? What pairs of tissues are you comparing? Spearman correlation is reported instead of Pearson, which is the primary metric used throughout the text.
The dPSI+ and dPSI- sets were indeed not well defined in the original submission. Moreover, we found our own code lacked consistency due to different tests executed at different times/by different people. We apologize for this lack of consistency and clarity which we worked to remedy in the revised version. To answer the reviewer’s question, given two tissues ($c,c'$), dPSI+ and dPSI- is for correctly classifying the exons that are significantly differentially included or excluded. Specifically, differential included exons are those for which $\Delta \Psi_{e,c1,c2} = \Psi_\Psi_{e,c1} - \Psi_{e,c2} \geq 0.15$, compared to those that are not ($\Delta \Psi_{e,c1,c2} < 0.05). Similarly, dPSI- is for correctly classifying the exons that are significantly differentially excluded in the first tissue or included in the second tissue ($\Delta \Psi_{e,c1,c2} = \Psi_\Psi_{e,c1} - \Psi_{e,c2} \leq -0.15$) compared to those that are not ($\Delta \Psi_{e,c1,c2} > -0.05). This means dPSI+ and dPSI- are dependent on the order of c1, c2. In addition, we also define a direction/order agnostic test for changing vs non changing events i.e. $|\Delta \Psi_{e,c1,c2}| \geq 0.15$ vs $|\Delta \Psi_{e,c1,c2}| < 0.05$. These test definitions are consistent with previous publications (e.g. Barash et al Nature 2010, Jha et al 2017) and also answer different biological questions: For example “Exons that go up in brain” and “Exons that go up in Liver” can reflect distinct mechanisms, while changing exons capture a model’s ability to identify regulated exons even if the direction of prediction may be wrong. The updated Appendix Figure 2 is now in the main text as Figure 2d and uses Pearson, while AUPRC and AUROC refer to the changing vs no-changing classification task described above such that we avoid dPSI+ and dPSI- when summarizing in this table over 3 pairs of tissues . Finally, we note that making sure all tests comply with the above definition also resulted in an update to Figure 2b/c labels and values, where TrASPr’s improvements over Pangolin reaches up to 1.8fold in AUPRC compared to 2.4fold in the earlier version. We again apologize for having a lack of clarity and consistent evaluations in the original submission.
(20) Minor typographical comments:
- Some plots could use more polishing (e.g., thicker stroke, bigger font size, consistent style (compare 4a to the other plots)...).
Agreed. While not critical for the science itself we worked to improve figure polishing in the revision to make those more readable and pleasant.
- Consider using 2-dimensional histograms instead of the current kernel density plots, which tend to over-smooth the data and hide potentially important details.
We were not sure what the exact suggestion is here and opted to leave the plots as is.
- L53: dPSI_{e, c, c'} is never formally defined. Is it PSI_{e, c} - PSI_{e, c'} or vice versa?
Definition now included (see above).
- L91: Define/explain "transformer" and provide reference.
We added the explanation and related reference of the transformer in the introduction section and BERT in the method section.
- L94: exons are short. Are you referring here to the flanking introns? Please explain.
We apologize for the lack of clarity. We are referring to a cassette exon alternative splicing event as is commonly defined by the splice junctions involved that is from the 5’ SS of the upstream exon to the 3’ SS of the downstream exon. The text now reads:
“...In contrast, 24% of the cassette exons analyzed in this study span a region between the flanking exons' upstream 3' and downstream 5' splice sites that are larger than 10 kb.”
- L132: It's unclear whether a single, shared transformer or four different transformers (one for each splice site) are being pre-trained. One would at least expect 5' and 3' splice sites to have a different transformer. In Methods, L506, it seems that each transformer is pre-trained separately.
We updated the text to read:
“We then center a dedicated transformer around each of the splice sites of the cassette exon and its upstream and downstream (competing) exons (four separate transformers for four splice sites in total).”
- L471: You explain here that it is unclear what tasks 'foundation' models are good for. Also in L128, you explain that you are not using a 'foundation' model. But then in L492, you describe the BERT model you're using as a foundation model!
Line 492 was simply a poor choice of wording as “foundation” is meant here simply as the “base component”. We changed it accordingly.
- L169, "pre-training ... BERT", explain what exactly this means. Is it using masking? Is it self-supervised learning? How many splice sites do you provide? Also explain more about the BERT architecture and provide references.
We added more details about the BERT architecture and training in the Methods section.
- L186 and later, the values for a and r provided here and in the below do not correspond to what is shown in Figure 2.
Fixed, thank you for noticing this.
- L187,188: What exactly do you mean by "events" and "samples"? Are they the same thing? If so, are they (exon, tissue) pairs? Please use consistent terminology. Moreover, when you say "changing between two conditions": do you take all six tissues whenever there is a 0.15 spread in PSI among them? Or do you take just the smallest PSI tissue and the largest PSI tissue when there is a 0.15 spread between them? Or something else altogether?
Reviewer #2 is yet again correct that the definitions were not precise. A “sample” involves a specific exon skipping “event” measured in two tissues. The text now reads:
“....most cassette exons do not change between a given tissue pair (only 14.0% of the samples in the dataset, i.e., a cassette exon measured across two tissues, exhibit |∆Ψ| ≥ 0.15). Thus, when we repeat this analysis only for samples involving exons that exhibited a change in inclusion (|∆Ψ| ≥ 0.15) between at least two tissues, performance degrades for all three models, but the differences between them become more striking (Figure 2a, right column).”
- Figure 1a, explain the colors in the figure legend. The 3D effect is not needed and is confusing (ditto in panel C).
Color explanation is now added: “exons and introns are shown as blue rectangles and black lines. The blue dashed line indicates the inclusive pattern and the red junction indicates an alternative splicing pattern.”
These are not 3D effects but stacks to indicate multiple events/cases. We agree these are not needed in Fig1a to illustrate types of AS and removed those. However, in Fig1c and matching caption we use the stacks to indicate HT data captures many such LSVs over which ML algorithms can be trained.
- Figure 1b, this cartoon seems unnecessary and gives the wrong impression that this paper explores mechanistic aspects of splicing. The only relevant fact (RBPs serving as splicing factors) can be explained in the text (and is anyway not really shown in this figure).
We removed Figure 1b cartoon.
- Figure 1c, what is being shown by the exon label "8"?
This was meant to convey exon ID, now removed to simplify the figure.
- Figure 1e, left, write "Intron Len" in one line. What features are included under "..."? Based on the text, I did not expect more features.
Also, the arrows emanating from the features do not make sense. Is "Embedding" a layer? I don't think so. Do not show it as a thin stripe. Finally, what are dPSI'+ and dPSI'-? are those separate outputs? are those logits of a classification task?
We agree this description was not good and have updated it in the revised version.
- Figure 1e, the right-hand side should go to a separate figure much later, when you introduce BOS.
We appreciate the suggestion. However, we feel that Figure 1e serves as a visual representation of the entire framework. Just like we opted to not turn this work into two separate papers (though we fully agree it is a valid option that would also increase our publication count), we also prefer to leave this unified visual representation as is.
- Figure 2, does the n=2456 refer to the number of (exons, tissues) pairs? So each exon contributes potentially six times to this plot? Typo "approximately".
The “n” refers to the number of samples which is a cassette event measured in two tissues. The same cassette event may appear in multiple samples if it was confidently quantified in more than two tissues. We updated the caption to reflect this and corrected the typo.
- Figure 2b, typo "differentially included (dPSI+) or excluded" .
Fixed.
- L221, "the DNABERT" => "DNABERT".
Fixed.
- L232, missing percent sign.
-
Fixed.
- L246, "see Appendix Section 2 for details" seems to instead refer to the third section of the appendix.
We do not have this as an Appendix, the reference has been updated.
- Figure 3, bottom panels, PSI should be "splice site usage"?
PSI is correct here - we hope the revised text/definitions make it more clear now.
- Figure 3b: typo: "when applied to alternative alternative 3'".
Fixed.
- p252, "polypyrimidine" (no capitalization).
Fixed.
- Strange capitalization of tissue names (e.g., "Brain-Cerebellum"). The tissue is called "cerebellum" without capitalization.
We used EBV (capital) for the abbreviation and lower case for the rest.
- Figure 4c: "predicted usage" on the left but "predicted PSI" on the right.
Right. We opted to leave it as is since Pangolin and SpliceAI do predict their definition of “usage” and not directly PSI, we just measure correlations to observed PSI as many works have done in the past.
- Figure 4 legend typo: "two three".
Fixed.
- L351, typo: "an (unsupervised)" (and no need to capitalize Transformer).
Fixed.
- L384, "compared to other tissues at least" => "compared to other tissues of at least".
Fixed.
- L549, P(Z) and P(S) are not defined in the text.
Fixed.
- L572, remove "Subsequently". Add missing citations at the end of the paragraph.
Fixed.
- L580-581, citations missing.
Fixed.
- L584-585, typo: "high confidince predictions"
Fixed.
- L659-660, BW-M and B-WM are both used. Typo?
Fixed.
- L895, "calculating the average of these two", not clear; please rewrite.
Fixed.
- L897, "Transformer" and "BERT", do these refer to the same thing? Be consistent.
BOS is a transformer and not a BERT but TrASPr uses the BERT architecture. BERT is a type of transformer as the reviewer is surely well aware so the sentence is correct. Still, to follow the reviewer’s recommendation for consistency/clarity we changed it here to state BERT.
- Appendix Figure 5: The term dPSI appears to be overloaded to also represent the difference between predicted PSI and measured PSI, which is inconsistent with previous definitions.
Indeed! We thank the reviewer again for their sharp eye and attention to details that we missed. We changed Supp Figure 5, now Figure 4 Supplementary Figure 2, to |PSI’-PSI| and defined those as the difference between TrASPr’s predictions (PSI’) and MAJIQ based PSI quantifications.
-
-
www.biorxiv.org www.biorxiv.org
-
Joint Public Review:
Summary:
This is an excellent, timely study investigating and characterizing the underlying neural activity that generates the neuroendocrine GnRH and LH surges that are responsible for triggering ovulation. Abundant evidence accumulated over the past 20 years implicated the population of kisspeptin neurons in the hypothalamic RP3V region (also referred to as the POA or AVPV/PeN kisspeptin neurons) as being involved in driving the GnRH surge in response to elevated estradiol (E2), also known as the "estrogen positive feedback". However, while former studies used Cfos coexpression as a marker of RP3V kisspeptin neuron activation at specific times and found this correlates with the timing of the LH surge, detailed examination of the live in vivo activity of these neurons before, during, and after the LH surge remained elusive due to technical challenges.
Here, Zhou and colleagues use fiber photometry to measure the long-term synchronous activity of RP3V kisspeptin neurons across different stages of the mouse estrous cycle, including on proestrus when the LH surge occurs, as well as in a well-established OVX+E2 mouse model of the LH surge.
The authors report that RP3V kisspeptin neuron activity is low on estrous and diestrus, but increases on proestrus several hours before the late afternoon LH surge, mirroring prior reports of rising GnRH neuron activity in proestrus female mice. The measured increase in RP3V kisspeptin activation is long, spanning ~13 hours in proestrus females and extending well beyond the end of the LH secretion, and is shown by the authors to be E2 dependent.
For this work, Kiss-Cre female mice received a Cre-dependent AAV injection, containing GCaMP6, to measure the neuronal activation of RP3V Kiss1 cells. Females exhibited periods of increased neuronal activation on the day of proestrus, beginning several hours prior to the LH surge and lasting for about 12 hours. Though oscillations in the pattern of GCaMP fluorescence were occasionally observed throughout the ovarian cycle, the frequency, duration, and amplitude of these oscillations were significantly higher on the day of proestrus. This increase in RP3V Kiss1 neuronal activation that precedes the increase in LH supports the hypothesis that these neurons are critical in regulating the LH surge. The authors compare this data to new data showing a similar increased activation pattern in GnRH neurons just prior to the LH surge, further supporting the hypothesis that RP3V Kiss1 cell activation causes the release of kisspeptin to stimulate GnRH neurons and produce the LH surge.
Strengths:
This study provides compelling data demonstrating that RP3V kisspeptin neuronal activity changes throughout the ovarian cycle, likely in response to changes in estradiol levels, and that neuronal activation increases on the day of the LH surge.
The observed increase in RP3V kisspeptin neuronal activation precedes the LH surge, which lends support to the hypothesis that these neurons play a role in regulating the estradiol-induced LH surge. Continuing to examine the complexities of the LH surge and the neuronal populations involved, as done in this study, is critical for developing therapeutic treatments for women's reproductive disorders.
This innovative study uses a within-subject design to examine neuronal activation in vivo across multiple hormone milieus, providing a thorough examination of the changes in activation of these neurons. The variability in neuronal activity surrounding the LH surge across ovarian cycles in the same animals is interesting and could not be achieved without this within-subjects design. The inclusion and comparison of ovary-intact females and OVX+E2 females is valuable to help test mechanisms under these two valuable LH surge conditions, and allows for further future studies to tease apart minor differences in the LH surge pattern between these 2 conditions.
This study provides an excellent experimental setup able to monitor the daily activity of preoptic kisspeptin neurons in freely moving female mice. It will be a valuable tool to assess the putative role of these kisspeptin neurons in various aspects of altered female fertility (aging, pathologies...). This approach also offers novel and useful insights into the impact of E2 and circadian cues on the electrical activity of RP3V kisspeptin neurons.
An intriguing cyclical oscillation in kisspeptin neural activity every 90 minutes exists, which may offer critical insight into how the RP3V kisspeptin system operates. Interestingly, there was also variability in the onset and duration of RP3V Kisspeptin neuron activity between and within mice in naturally cycling females. Preoptic kisspeptin neurons show an increased activity around the light/dark transition only on the day of proestrus, and this is associated with an increase in LH secretion. An original finding is the observation that the peak of kisspeptin neuron activation continues a few hours past the peak of LH, and the authors hypothesize that this prolonged activity could drive female sexual behaviors, which usually appear after the LH surge.
The authors demonstrated that ovariectomy resulted in very little neuronal activity in RP3V kisspeptin neurons. When these ovarietomized females were treated with estradiol benzoate (EB) and an LH surge was induced, there was an increase in RP3V kisspeptin neuronal activation, as was seen during proestrus. However, the magnitude of the change in activity was greater during proestrus than during the EB-induced LH surge. Interestingly, the authors noted a consistent peak in activity about 90 minutes prior to lights out on each day of the ovarian cycle and during EB treatment, but not in ovariectomized females. The functional purpose of this consistent neuronal activity at this time remains to be determined.
Though not part of this study, the comparison of neuronal activation of GnRH neurons during the LH surge to the current data was convincing, demonstrating a similar pattern of increased activation that precedes the LH surge.
In summary, the study is well-designed, uses proper controls and analyses, has robust data, and the paper is nicely organized and written. The data from these experiments is compelling, and the authors' claims and conclusions are nicely supported and justified by the data. The data support the hypothesis in the field that these RP3V neurons regulate the LH surge. Overall, these findings are important and novel, and lend valuable insight into the underlying neural mechanisms for neuroendocrine control of ovulation.
Weaknesses:
(1) LH levels were not measured in many mice or in robust temporal detail, such as every 30 or 60 min, to allow a more detailed comparison between the fine-scale timing of RP3V neuron activation with onset and timing of LH surge dynamics.
(2) The authors report that the peak LH value occurred 3.5 hours after the first RP3V kisspeptin neuron oscillation. However, it is likely, and indeed evident from the 2 example LH patterns shown in Figures 3A-B, that LH values start to increase several hours before the peak LH. This earlier rise in LH levels ("onset" of the surge) occurs much closer in time to the first RP3V kisspeptin neuron oscillatory activation, and as such, the ensuing LH secretion may not be as delayed as the authors suggest.
(3) The authors nicely show that there is some variation (~2 hours) in the peak of the first oscillation in proestrus females. Was this same variability present in OVX+E2 females, or was the variability smaller or absent in OVX+E2 versus proestrus? It is possible that the variability in proestrus mice is due to variability in the timing and magnitude of rising E2 levels, which would, in theory, be more tightly controlled and similar among mice in the OVX+E2 model. If so, the OVX+E2 mice may have less variability between mice for the onset of RP3V kisspeptin activity.
(4) One concern regarding this study is the lack of data showing the specificity of the AAV and the GCaMP6s signals. There are no data showing that GCaMP6s is limited to the RP3V and is not expressed in other Kiss1 populations in the brain. Given that 2ul of the AAV was injected, which seems like a lot considering it was close to the ventricle, it is important to show that the signal and measured activity are specific to the RP3V region. Though the authors discuss potential reasons for the low co-expression of GCaMP6 and kisspeptin immunoreactivity, it does raise some concern regarding the interpretation of these results. The low co-expression makes it difficult to confirm the Kiss1 cell-specificity of the Cre-dependent AAV injections. In addition, if GFP (GCaMP6s) and kisspeptin protein co-localization is low, it is possible that the activation of these neurons does not coincide with changes in kisspeptin or that these neurons are even expressing Kiss1 or kisspeptin at the time of activation. It is important to remember that the study measures activation of the kisspeptin neuron, and it does not reveal anything specific about the activity of the kisspeptin protein.
(5) One additional minor concern is that LH levels were not measured in the ovariectomized females during the expected time of the LH surge. The authors suggest that the lower magnitude of activation during the LH surge in these females, in comparison to proestrus females, may be the result of lower LH levels. It's hard to interpret the difference in magnitude of neuronal activation between EB-treated and proestrus females without knowing LH levels. In addition, it's possible that an LH surge did not occur in all EB-treated females, and thus, having LH levels would confirm the success of the EB treatment.
(6) This kisspeptin neuron peak activity is abolished in ovariectomized mice, and estradiol replacement restored this activity, but only partially. Circulating levels of estradiol were not measured in these different setups, but the authors hypothesize that the lack of full restoration may be due to the absence of other ovarian signals, possibly progesterone.
(7) Recordings in several mice show inter- and intra-variability in the time of peak onset. It is not shown whether this variability is associated with a similar variability in the timing of the LH surge onset in the recorded mice. The authors hypothesized that this variability indicates a poor involvement of the circadian input. However, no experiments were done to investigate the role of the (vasopressinergic-driven) circadian input on the kisspeptin neuron activation at the light/dark transition. Thus, we suggest that the authors be more tentative about this hypothesis.
-
-
www.youtube.com www.youtube.com
-
12:48 "I have a hypothesis about why society is broken right now ... 1913 fiat currency ... printing money ... inflation ... steal from people ... hyper inequality ... 10% of Americans own 93% of assets ... unfair ... envy ... hollowing out of the middle class ... wealth inequality is reaching levels like in the French revolution ... we actually are headed towards a French revolution, but it's economic in nature."
no. fiat money is just another tool to control idiots, just like religion or politics. the actual drivers of this global policy are: overpopulation, degeneration, ecocide, resource depletion.
so yes, the problem is "economic in nature" in the sense that we have too many humans and too few planets.
the obvious solution is global depopulation by 90% of today's population, but that is obviously not a popular topic, because only 10% will survive this cull.
-
-
uniworldstudios.com uniworldstudios.com
-
When identical keywords exist across multiple ad groups, your ads bid against each other in Google’s mini-auctions. This means you’re not just competing with other advertisers—you’re competing with yourself, needlessly inflating costs. Over time, this internal bidding war can lead to wasted budget allocations, especially if high-volume keywords are involved. Each bid siphons away funds that could have been allocated to more strategic, unique campaigns, ultimately diminishing your ROI.
They aren't LITERALLY competing against each other in the auction. It's not that each is setting a bid and then trying to one up each other. It's that if one KW has weaker relevance it might bid really high, resulting in a higher ad rank & winning the same click for a more expensive price, just because the structure wasn't clean enough. We also run the risk of Google favoring click heavy, but low CvR ad ad group placements.
-
-
borretti.me borretti.me
-
I wish people's works were more online—more accessible (preferably, with stable, citable URLs). But to say that the people themselves should be more public in the sense that the author means here is just a continuation-without-lessons-learned of the modern (Facebook and onwards) social networking era.
It's great if all of a person's contributions to, say, mailing lists are preserved and available—and aren't just holes, missing from the record.* That's different from whether or not it's great to be able to click on that person's name, find a profile page for them, and then encounter an exhaustive, reverse chronological feed of all of their activity across all mailing lists. Mailing lists rarely enable this, but virtually every modern social network does, and they're actually built around it.
The former is topic-based indexing, and the latter is actor- (person-) based indexing. Actor-based indexing is bad, and we know that it's bad.
Actor-based indexing is like running into someone you know (or a stranger, even) at the post office and then, through some mechanism where their physical presence wired to some metaverse data source, being able to perform some tap/gesture at the virtualized floating bubble over their heads that lets you see all the public places where you would have seen them earlier that day if you had been at any one of those, and then having a log of every interaction for the day prior, and the day before that, and so on, stretching back over their entire life, including the grocery store, the restaurant, the houseparty they were at, their work, etc. This would be bad. That means it's not good. And it's not good "online", either, for the exact same reasons.
* as unfortunately, many Mastodon (and other ActivityPub-powered) interactions turn out; Mastodon happens to achieve the worst of both worlds!
-
-
www.ctol.digital www.ctol.digital
-
That raises fixed costs and favors scale—paradoxically advantaging the largest platforms that can afford regional bifurcation while crushing subscale competitors.
Not really, it does not favor scale, as it's progressive compliance. The next sentence says as much. One platform's bifurcation is the same as having two smaller ones, who have less compliance costs and thus won't be crushed. Which is already the case even, global platforms already cater to diff regulatory regimes (and morally questionable at that). The underlying faulty assumption is that of global platforms for everyone and everything being the desired outcome at all. SV thinking and funding is the root cause. Other paths exist, just not in their world. Zebra's not unicorns. [[Zebra bedrijven zijn beter 20190907063530]] Vgl physical e.g. the German industrial base actually is one of many medium sized orgs being market leaders in some niche, not the car manufacturers usually mentioned as such.
-
-
www.niemanlab.org www.niemanlab.org
-
The cost goes beyond simple inefficiency and becomes a mountain of invisible labor, usually absorbed by the most junior person in the room or whoever has the misfortune of being labeled as “good with computers.” It becomes a drag on every collaboration, the friction in every workflow, the meetings that take an extra ten minutes while someone (who is often paid twice the average salary of the other people in the meeting) figures out why they can’t access the shared folder the rest of us have been using for months. It’s the quiet erosion of patience and goodwill among people who are constantly expected to know and fix things that shouldn’t need fixing in the first place.
The cost of lack of skills is not just in the individual knowledge worker, it gets externalised to others to fix it, or multiplied in groups waiting on you to get something working. The incompetence spreads out.
-
-
www.biorxiv.org www.biorxiv.org
-
Note: This response was posted by the corresponding author to Review Commons. The content has not been altered except for formatting.
Learn more at Review Commons
Reply to the reviewers
Reviewer #1* (Evidence, reproducibility and clarity (Required)):
Summary: In this study, the authors used proximity proteomics in U2OS cells to identify several E3 ubiquitin ligases recruited to stress granules (SGs), and they focused on MKRN2 as a novel regulator. They show that MKRN2 localization to SGs requires active ubiquitination via UBA1. Functional experiments demonstrated that MKRN2 knockdown increases the number of SG condensates, reduces their size, slightly raises SG liquidity during assembly, and slows disassembly after heat shock. Overexpression of MKRN2-GFP combined with confocal imaging revealed co-localization of MKRN2 and ubiquitin in SGs. By perturbing ubiquitination (using a UBA1 inhibitor) and inducing defective ribosomal products (DRiPs) with O-propargyl puromycin, they found that both ubiquitination inhibition and MKRN2 depletion lead to increased accumulation of DRiPs in SGs. The authors conclude that MKRN2 supports granulostasis, the maintenance of SG homeostasis , through its ubiquitin ligase activity, preventing pathological DRiP accumulation within SGs.
Major comments: - Are the key conclusions convincing? The key conclusions are partially convincing. The data supporting the role of ubiquitination and MKRN2 in regulating SG condensate dynamics are coherent, well controlled, and consistent with previous literature, making this part of the study solid and credible. However, the conclusions regarding the ubiquitin-dependent recruitment of MKRN2 to SGs, its relationship with UBA1 activity, the functional impact of the MKRN2 knockdown for DRiP accumulation are less thoroughly supported. These aspects would benefit from additional mechanistic evidence, validation in complementary model systems, or the use of alternative methodological approaches to strengthen the causal connections drawn by the authors. - Should the authors qualify some of their claims as preliminary or speculative, or remove them altogether? The authors should qualify some of their claims as preliminary. 1) MKRN2 recruitment to SGs (ubiquitin-dependent): The proteomics and IF data are a reasonable starting point, but they do not yet establish that MKRN2 is recruited from its physiological localization to SGs in a ubiquitin-dependent manner. To avoid overstating this point the authors should qualify the claim and/or provide additional controls: show baseline localization of endogenous MKRN2 under non-stress conditions (which is reported in literature to be nuclear and cytoplasmatic), include quantification of nuclear/cytoplasmic distribution, and demonstrate a shift into bona fide SG compartments after heat shock. Moreover, co-localization of overexpressed GFP-MKRN2 with poly-Ub (FK2) should be compared to a non-stress control and to UBA1-inhibition conditions to support claims of stress- and ubiquitination-dependent recruitment. *
Authors: We will stain cells for endogenous MKRN2 and quantify nuc/cyto ratio of MKRN2 without heat stress, without heat stress + TAK243, with HS and with HS + TAK243. We will do the same in the MKRN2-GFP overexpressing line while also staining for FK2.
*2) Use and interpretation of UBA1 inhibition: UBA1 inhibition effectively blocks ubiquitination globally, but it is non-selective. The manuscript should explicitly acknowledge this limitation when interpreting results from both proteomics and functional assays. Proteomics hits identified under UBA1 inhibition should be discussed as UBA1-dependent associations rather than as evidence for specific E3 ligase recruitment. The authors should consider orthogonal approaches before concluding specificity. *
Authors: We have acknowledged the limitation of using only TAK243 in our study by rephrasing statements about dependency on “ubiquitination” to “UBA1-dependent associations”.
* 3) DRiP accumulation and imaging quality: The evidence presented in Figure 5 is sufficient to substantiate the claim that DRiPs accumulate in SGs upon ubiquitination inhibition or MKRN2 depletion but to show that the event of the SGs localization and their clearance from SGs during stress is promoted by MKRN3 ubiquitin ligase activity more experiments would be needed. *
Authors: We have acknowledged the fact that our experiments do not include DRiP and SG dynamics assays using ligase-dead mutants of MKRN2 by altering our statement regarding MKRN2-mediated ubiquitination of DRiPs in the text (as proposed by reviewer 1).
*- Would additional experiments be essential to support the claims of the paper? Request additional experiments only where necessary for the paper as it is, and do not ask authors to open new lines of experimentation. Yes, a few targeted experiments would strengthen the conclusions without requiring the authors to open new lines of investigation. 1) Baseline localization of MKRN2: It would be important to show the baseline localization of endogenous and over-expressed MKRN2 (nuclear and cytoplasmic) under non-stress conditions and prior to ubiquitination inhibition. This would provide a reference to quantify redistribution into SGs and demonstrate recruitment in response to heat stress or ubiquitination-dependent mechanisms. *
Authors: We thank the reviewer for bringing this important control. We will address it in revisions.
We will quantify the nuclear/cytoplasmic distribution of endogenous and GFP-MKRN2 under control, TAK243, heat shock, and combined conditions, and assess MKRN2–ubiquitin colocalization by FK2 staining in unstressed cells.
* 2) Specificity of MKRN2 ubiquitin ligase activity: to address the non-specific effects of UBA1 inhibition and validate that observed phenotypes depend on MKRN2's ligase activity, the authors could employ a catalytically inactive MKRN2 mutant in rescue experiments. Comparing wild-type and catalytic-dead MKRN2 in the knockdown background would clarify the causal role of MKRN2 activity in SG dynamics and DRiP clearance. *
Authors: We thank the reviewer for this suggestion and have altered the phrasing of some of our statements in the text accordingly.
* 3) Ubiquitination linkage and SG marker levels: While the specific ubiquitin linkage type remains unknown, examining whether MKRN2 knockdown or overexpression affects total levels of key SG marker proteins would be informative. This could be done via Western blotting of SG markers along with ubiquitin staining, to assess whether MKRN2 influences protein stability or turnover through degradative or non-degradative ubiquitination. Such data would strengthen the mechanistic interpretation while remaining within the current study's scope. *
Authors: We thank the reviewer for requesting and will address it by performing MKRN2 KD and perform Western blot for G3BP1.
*
- Are the suggested experiments realistic in terms of time and resources? It would help if you could add an estimated cost and time investment for substantial experiments. The experiments suggested in points 1 and 3 are realistic and should not require substantial additional resources beyond those already used in the study. • Point 1 (baseline localization of MKRN2): This involves adding two control conditions (no stress and no ubiquitination inhibition) for microscopy imaging. The setup is essentially the same as in the current experiments, with time requirements mainly dependent on cell culture growth and imaging. Overall, this could be completed within a few weeks. • Point 3 (SG marker levels and ubiquitination): This entails repeating the existing experiment and adding a Western blot for SG markers and ubiquitin. The lab should already have the necessary antibodies, and the experiment could reasonably be performed within a couple of weeks. • Point 2 (catalytically inactive MKRN2 mutant and rescue experiments): This is likely more time-consuming. Designing an effective catalytic-dead mutant depends on structural knowledge of MKRN2 and may require additional validation to confirm loss of catalytic activity. If this expertise is not already present in the lab, it could significantly extend the timeline. Therefore, this experiment should be considered only if similarly recommended by other reviewers, as it represents a higher resource and time investment.
Overall, points 1 and 3 are highly feasible, while point 2 is more substantial and may require careful planning.
-
Are the data and the methods presented in such a way that they can be reproduced? Yes. The methodologies used in this study to analyze SG dynamics and DRiP accumulation are well-established in the field and should be reproducible, particularly by researchers experienced in stress granule biology. Techniques such as SG assembly and disassembly assays, use of G3BP1 markers, and UBA1 inhibition are standard and clearly described. The data are generally presented in a reproducible manner; however, as noted above, some results would benefit from additional controls or complementary experiments to fully support specific conclusions.
-
Are the experiments adequately replicated and statistical analysis adequate? Overall, the experiments in the manuscript appear to be adequately replicated, with most assays repeated between three and five times, as indicated in the supplementary materials. The statistical analyses used are appropriate and correctly applied to the datasets presented. However, for Figure 5 the number of experimental replicates is not reported. This should be clarified, and if the experiment was not repeated sufficiently, additional biological replicates should be performed. Given that this figure provides central evidence supporting the conclusion that DRiP accumulation depends on ubiquitination-and partly on MKRN2's ubiquitin ligase activity-adequate replication is essential. *
Authors: We thank the reviewer for noting this accidental omission. We now clarify in the legend of Figure 5 that the experiments with DRiPs were replicated three times.
Minor comments: - Specific experimental issues that are easily addressable. • For the generation and the validation of MKRN2 knockdown in UOS2 cells data are not presented in the results or in the methods sections to demonstrate the effective knockdown of the protein of interest. This point is quite essential to demonstrate the validity of the system used
Authors: We thank the reviewer for requesting and will address it by performing MKRN2 KD and perform Western blot and RT-qPCR.
-
* In the supplementary figure 2 it would be useful to mention if the Western Blot represent the input (total cell lysates) before the APEX-pulldown or if it is the APEX-pulldown loaded for WB. There is no consistence in the difference of biotynilation between different replicates shown in the 2 blots. For example in R1 and R2 G3BP1-APX TAK243 the biotynilation is one if the strongest condition while on the left blot, in the same condition comparison samples R3 and R4 are less biotinilated compared to others. It would be useful to provide an explanation for that to avoid any confusion for the readers. * Authors: We have added a mention in the legend of Figure S2 that these are total cell lysates before pulldown. The apparent differences in biotin staining are small and not sufficient to question the results of our APEX-proteomics.
-
* In Figure 2D, endogenous MKRN2 localization to SGs appears reduced following UBA1 inhibition. However, it is not clear whether this reduction reflects a true relocalization or a decrease in total MKRN2 protein levels. To support the interpretation that UBA1 inhibition specifically affects MKRN2 recruitment to SGs rather than its overall expression, the authors should provide data showing total MKRN2 levels remain unchanged under UBA1 inhibition, for example via Western blot of total cell lysates. * Authors: Based on first principles in regulation of gene expression, it is unlikely that total MKRN2 expression levels would decrease appreciably through transcriptional or translational regulation within the short timescale of these experiments (1 h TAK243 pretreatment followed by 90 min of heat stress).
-
* DRIPs accumulation is followed during assembly but in the introduction is highlighted the fact that ubiquitination events, other reported E3 ligases and in this study data on MKRN2 showed that they play a crucial role in the disassembly of SGs which is also related with cleareance of DRIPs. Authors could add tracking DRIPs accumulation during disassembly to be added to Figure 5. I am not sure about the timeline required for this but I am just adding as optional if could be addressed easily. * Authors: We thank the reviewer for proposing this experimental direction. However, in a previous study (Ganassi et al., 2016; 10.1016/j.molcel.2016.07.021), we demonstrated that DRiP accumulation during the stress granule assembly phase drives conversion to a solid-like state and delays stress granule disassembly. It is therefore critical to assess DRiP enrichment within stress granules immediately after their formation, rather than during the stress recovery phase, as done here.
-
* The authors should clarify in the text why the cutoff used for the quantification in Figure 5D (PC > 3) differs from the cutoff used elsewhere in the paper (PC > 1.5). Providing a rationale for this choice will help the reader understand the methodological consistency and ensure that differences in thresholds do not confound interpretation of the results. * Authors: We thank the reviewer for this question. The population of SGs with a DRiP enrichment > 1.5 represents SGs with a significant DRiP enrichment compared to the surrounding (background) signal. As explained in the methods, the intensity of DRiPs inside each SG is corrected by the intensity of DRiPs two pixels outside of each SG. Thus, differences in thresholds between independent experimental conditions (5B versus 5D) do not confound interpretation of the results but depend on overall staining intensity that can different between different experimental conditions. Choosing the cut-off > 3 allows to specifically highlight the population of SGs that are strongly enriched with DRiPs. MKRN2 silencing caused a strong DRiP enrichment in the majority of the SGs analyzed and therefore we chose this way of data representation. Note that the results represent the average of the analysis of 3 independent experiments with high numbers of SGs automatically segmented and analyzed/experiment. Figure 5A, B: n = 3 independent experiments; number of SGs analyzed per experiment: HS + OP-puro (695; 1216; 952); TAK-243 + HS + OP-puro (1852; 2214; 1774). Figure 5C, D: n = 3 independent experiments; number of SGs analyzed per experiment: siRNA control, HS + OP-puro (1984; 1400; 1708); siRNA MKRN2, HS + OP-puro (912; 1074; 1532).
-
* For Figure 3G, the authors use over-expressed MKRN2-GFP to assess co-localization with ubiquitin in SGs. Given that a reliable antibody for endogenous MKRN2 is available and that a validated MKRN2 knockdown line exists as an appropriate control, this experiment would gain significantly in robustness and interpretability if co-localization were demonstrated using endogenous MKRN2. In the current over-expression system, MKRN2-GFP is also present in the nucleus, whereas the endogenous protein does not appear nuclear under the conditions shown. This discrepancy raises concerns about potential over-expression artifacts or mislocalization. Demonstrating co-localization using endogenous MKRN2 would avoid confounding effects associated with over-expression. If feasible, this would be a relatively straightforward experiment to implement, as it relies on tools (antibody and knockdown line) already described in the manuscript.
* Authors: We thank the reviewer for requesting and will address it by performing MKRN2 KD, FK2 immunofluorescence microscopy and perform SG partition coefficient analysis.
* - Are prior studies referenced appropriately? • From line 54 to line 67, the manuscript in total cites eight papers regarding the role of ubiquitination in SG disassembly. However, given the use of UBA1 inhibition in the initial MS-APEX experiment and the extensive prior literature on ubiquitination in SG assembly and disassembly under various stress conditions, the manuscript would benefit from citing additional relevant studies to provide more specifc examples. Expanding the references would provide stronger context, better connect the current findings to prior work, and emphasize the significance of the study in relation to established literature *
Authors: We have added citations for the relevant studies.
- *
At line 59, it would be helpful to note that G3BP1 is ubiquitinated by TRIM21 through a Lys63-linked ubiquitin chain. This information provides important mechanistic context, suggesting that ubiquitination of SG proteins in these pathways is likely non-degradative and related to functional regulation of SG dynamics rather than protein turnover. * Authors: The reviewer is correct. We have added to the text that G3BP1 is ubiquitinated through a Lys63-linked ubiquitin chain.
- *
When citing references 16 and 17, which report that the E3 ligases TRIM21 and HECT regulate SG formation, the authors should provide a plausible explanation for why these specific E3 ligases were not detected in their proteomics experiments. Differences could arise from the stress stimulus used, cell type, or experimental conditions. Similarly, since MKRN2 and other E3 ligases identified in this study have not been reported in previous works, discussing these methodological or biological differences would help prevent readers from questioning the credibility of the findings. It would also be valuable to clarify in the Conclusion that different types of stress may activate distinct ubiquitination pathways, highlighting context-dependent regulation of SG assembly and disassembly. * Authors: We thank the reviewer for this suggestion. We added to the discussion plausible explanations for why our study identified new E3 ligases.
- *
Line 59-60: when referring to the HECT family of E3 ligases involved in ubiquitination and SG disassembly, it would be more precise to report the specific E3 ligase identified in the cited studies rather than only the class of ligase. This would provide clearer mechanistic context and improve accuracy for readers. * Authors: We have added this detail to the discussion.
- *
The specific statement on line 182 "SG E3 ligases that depend on UBA1 activity are RBULs" should be supported by reference. * Authors: We have added citations to back up our claim that ZNF598, CNOT4, MKRN2, TRIM25 and TRIM26 exhibit RNA-binding activity.
*- Are the text and figures clear and accurate?
• In Supplementary Figure 1, DMSO is shown in green and the treatment in red, whereas in the main figures (Figure 1B and 1F) the colours in the legend are inverted. To avoid confusion, the colour coding in figure legends should be consistent across all figures throughout the manuscript. *
Authors: We have made the colors consistent across the main and supplementary figures.
- *
At line 79, the manuscript states that "inhibition of ubiquitination delayed fluorescence recovery dynamics of G3BP1-mCherry, relative to HS-treated cells (Figure 1F, Supplementary Fig. 6A)." However, the data shown in Figure 1F appear to indicate the opposite effect: the TAK243-treated condition (green curve) shows a faster fluorescence recovery compared to the control (red curve). This discrepancy between the text and the figure should be corrected or clarified, as it may affect the interpretation of the role of ubiquitination in SG dynamics. * Authors: Good catch. We now fixed the graphical mistake (Figure 1F and S6).
-
* Line 86: adjust a missing bracket * Authors: Thank you, we fixed it.
-
*
There appears to be an error in the legend of Supplementary Figure 3: the legend states that the red condition (MKRN2) forms larger aggregates, but both the main Figure 3C of the confocal images and the text indicate that MKRN2 (red) forms smaller aggregates. Please correct the legend and any corresponding labels so they are consistent with the main figure and the text. The authors should also double-check that the figure panel order, color coding, and statistical annotations match the legend and the descriptions in the Results section to avoid reader confusion.
* Authors: This unfortunate graphical mistake has been corrected.
-
* At lines 129-130, the manuscript states that "FRAP analysis demonstrated that MKRN2 KD resulted in a slight increase in SG liquidity (Fig. 3F, Supplementary Fig. 6B)." However, the data shown in Figure 3F appear to indicate the opposite trend: the MKRN2 KD condition (red curve) exhibits a faster fluorescence recovery compared to the control (green curve). This discrepancy between the text and the figure should be corrected or clarified, as it directly affects the interpretation of MKRN2's role in SG disassembly. Ensuring consistency between the written description and the plotted FRAP data is essential for accurate interpretation. * Authors: We thank the reviewer and clarify in the legend of Figure 3F and the Results the correct labels: indeed faster fluorescence recovery seen in MKRN2 KD is correctly interpreted as increased liquidity in the text.
-
*
At lines 132-133, the manuscript states: "Then, to further test the impact of MKRN2 on SG dynamics, we overexpressed MKRN2-GFP and observed that it was recruited to SG (Fig. 3G)." This description should be corrected or clarified, as the over-expressed MKRN2-GFP also appears to localize to the nucleus. * Authors: The text has been modified to reflect both the study of MKRN2 localization to SGs and of nuclear localization.
- *
At lines 134-135, the manuscript states that the FK2 antibody detects "free ubiquitin." This is incorrect. FK2 does not detect free ubiquitin; it recognizes only ubiquitin conjugates, including mono-ubiquitinated and poly-ubiquitinated proteins. The text should be corrected accordingly to avoid misinterpretation of the immunostaining data. * Authors: Thank you for pointing out this error. We have corrected it.
- * Figure 5A suffers from poor resolution, and no scale bar is provided, which limits interpretability. Additionally, the ROI selected for the green channel (DRIPs) appears to capture unspecific background staining, while the most obvious DRIP spots are localized in the nucleus. The authors should clarify this in the text, improve the image quality if possible, and ensure that the ROI accurately represents DRIP accumulation - in SGs rather than background signal. * Authors: We thank the reviewer for pointing the sub-optimal presentation of this figure. We modified Figure 5A to improve image quality and interpretation. Concerning the comment that “the most obvious DRIP spots are localized in the nucleus”, this is in line with our previous findings demonstrating that a fraction of DRiPs accumulates in nucleoli (Mediani et al. 2019 10.15252/embj.2018101341). To avoid misinterpretation, we modified Figure 5A as follows: (i) we provide a different image for control cells, exposed to heat shock and OP-puro; (ii) we select a ROI that only shows a few stress granules; (iii) we added arrowheads to indicate the nucleoli that are strongly enriched for DRiPs; (iv) we include a dotted line to show the nuclear membrane, helping to distinguish cytoplasm and nucleus in the red and green channel. We also include the scale bars (5 µm) in the image.
* Do you have suggestions that would help the authors improve the presentation of their data and conclusions?
• In the first paragraph following the APEX proteomics results, the authors present validation data exclusively for MKRN2, justifying this early focus by stating that MKRN2 is the most SG-depleted E3 ligase. However, in the subsequent paragraph they introduce the RBULs and present knockdown data for MKRN2 along with two additional E3 ligases identified in the screen, before once again emphasizing that MKRN2 is the most SG-depleted ligase and therefore the main focus of the study. For clarity and logical flow, the manuscript would benefit from reordering the narrative. Specifically, the authors should first present the validation data for all three selected E3 ligases, and only then justify the decision to focus on MKRN2 for in-depth characterization. In addition to the extent of its SG depletion, the authors may also consider providing biologically relevant reasons for prioritizing MKRN2 (e.g., domain architecture, known roles in stress responses, or prior evidence of ubiquitination-related functions). Reorganizing this section would improve readability and better guide the reader through the rationale for the study's focus.*
Authors: We thank the reviewer for this suggested improvement to our “storyline”. As suggested by the reviewer, we have moved the IF validation of MKRN2 to the following paragraph in order to improve the flow of the manuscript. We added additional justification to prioritizing MKRN2 citing (Youn et al. 2018 and Markmiller et al. 2018).
- *
At lines 137-138, the manuscript states: "Together these data indicate that MKRN2 regulates the assembly dynamics of SGs by promoting their coalescence during HS and can increase SG ubiquitin content." While Figure 3G shows some co-localization of MKRN2 with ubiquitin, immunofluorescence alone is insufficient to claim an increase in SG ubiquitin content. This conclusion should be supported by orthogonal experiments, such as Western blotting, in vitro ubiquitination assays, or immunoprecipitation of SG components. Including a control under no-stress conditions would also help demonstrate that ubiquitination increases specifically in response to stress. The second part of the statement should therefore be rephrased to avoid overinterpretation, for example:"...and may be associated with increased ubiquitination within SGs, as suggested by co-localization, pending further validation by complementary assays." * Authors: The statement has been rephrased in a softer way as suggested by the reviewer.
-
At line 157, the statement: "Therefore, we conclude that MKRN2 ubiquitinates a subset of DRiPs, avoiding their accumulation inside SGs" should be rephrased as a preliminary observation. While the data support a role for MKRN2 in SG disassembly and a reduction of DRIPs, direct ubiquitination of DRIPs by MKRN2 has not been demonstrated. A more cautious phrasing would better reflect the current evidence and avoid overinterpretation. * * *Authors: We thank the reviewer for this suggestion and have altered the phrasing of this statement accordingly.
*Reviewer #1 (Significance (Required)):
General assessment: provide a summary of the strengths and limitations of the study. What are the strongest and most important aspects? What aspects of the study should be improved or could be developed?
• This study provides a valuable advancement in understanding the role of ubiquitination in stress granule (SG) dynamics and the clearance of SGs formed under heat stress. A major strength is the demonstration of how E3 ligases identified through proteomic screening, particularly MKRN2, influence SG assembly and disassembly in a ubiquitination- and heat stress-dependent manner. The combination of proteomics, imaging, and functional assays provides a coherent mechanistic framework linking ubiquitination to SG homeostasis. Limitations of the study include the exclusive use of a single model system (U2OS cells), which may limit generalizability. Additionally, some observations-such as MKRN2-dependent ubiquitination within SGs and changes in DRIP accumulation under different conditions-would benefit from orthogonal validation experiments (e.g., Western blotting, immunoprecipitation, or in vitro assays) to confirm and strengthen these findings. Addressing these points would enhance the robustness and broader applicability of the conclusions.
Advance: compare the study to the closest related results in the literature or highlight results reported for the first time to your knowledge; does the study extend the knowledge in the field and in which way? Describe the nature of the advance and the resulting insights (for example: conceptual, technical, clinical, mechanistic, functional,...).
• The closest related result in literature is - Yang, Cuiwei et al. "Stress granule homeostasis is modulated by TRIM21-mediated ubiquitination of G3BP1 and autophagy-dependent elimination of stress granules." Autophagy vol. 19,7 (2023): 1934-1951. doi:10.1080/15548627.2022.2164427 - demonstrating that TRIM21, an E3 ubiquitin ligase, catalyzes K63-linked ubiquitination of G3BP1, a core SG nucleator, under oxidative stress. This ubiquitination by TRIM21 inhibits SG formation, likely by altering G3BP1's propensity for phase separation. In contrast, the MKRN2 study identifies a different E3 (MKRN2) that regulates SG dynamics under heat stress and appears to influence both assembly and disassembly. This expands the role of ubiquitin ligases in SG regulation beyond those previously studied (like TRIM21).
• Gwon and colleagues (Gwon Y, Maxwell BA, Kolaitis RM, Zhang P, Kim HJ, Taylor JP. Ubiquitination of G3BP1 mediates stress granule disassembly in a context-specific manner. Science. 2021;372(6549):eabf6548. doi:10.1126/science.abf6548) have shown that K63-linked ubiquitination of G3BP1 is required for SG disassembly after heat stress. This ubiquitinated G3BP1 recruits the segregase VCP/p97, which helps extract G3BP1 from SGs for disassembly. The MKRN2 paper builds on this by linking UBA1-dependent ubiquitination and MKRN2's activity to SG disassembly. Specifically, they show MKRN2 knockdown affects disassembly, and suggest MKRN2 helps prevent accumulation of defective ribosomal products (DRiPs) in SGs, adding a new layer to the ubiquitin-VCP model.
• Ubiquitination's impact is highly stress- and context-dependent (different chain types, ubiquitin linkages, and recruitment of E3s). The MKRN2 work conceptually strengthens this idea: by showing that MKRN2's engagement with SGs depends on active ubiquitination via UBA1, and by demonstrating functional consequences (SG dynamics + DRIP accumulation), the study highlights how cellular context (e.g., heat stress) can recruit specific ubiquitin ligases to SGs and modulate their behavior.
• There is a gap in the literature: very few (if any) studies explicitly combine the biology of DRIPs, stress granules, and E3 ligase mediated ubiquitination, especially in mammalian cells. There are relevant works about DRIP biology in stress granules, but those studies focus on chaperone-based quality control, not ubiquitin ligase-mediated ubiquitination of DRIPs. This study seems to be one of the first to make that connection in mammalian (or human-like) SG biology. A work on the plant DRIP-E3 ligase TaSAP5 (Zhang N, Yin Y, Liu X, et al. The E3 Ligase TaSAP5 Alters Drought Stress Responses by Promoting the Degradation of DRIP Proteins. Plant Physiol. 2017;175(4):1878-1892. doi:10.1104/pp.17.01319 ) shows that DRIPs can be directly ubiquitinated by E3s in other biological systems - which supports the plausibility of the MKRN2 mechanism, but it's not the same context.
• A very recent review (Yuan, Lin et al. "Stress granules: emerging players in neurodegenerative diseases." Translational neurodegeneration vol. 14,1 22. 12 May. 2025, doi:10.1186/s40035-025-00482-9) summarizes and reinforces the relationship among SGs and the pathogenesis of different neurodegenerative diseases (NDDs). By identifying MKRN2 as a new ubiquitin regulator in SGs, the current study could have relevance for neurodegeneration and proteotoxic diseases, providing a new candidate to explore in disease models.
Audience: describe the type of audience ("specialized", "broad", "basic research", "translational/clinical", etc...) that will be interested or influenced by this research; how will this research be used by others; will it be of interest beyond the specific field?
The audience for this paper is primarily specialized, including researchers in stress granule biology, ubiquitin signaling, protein quality control, ribosome biology, and cellular stress responses. The findings will also be of interest to scientists working on granulostasis, nascent protein surveillance, and proteostasis mechanisms. Beyond these specific fields, the study provides preliminary evidence linking ubiquitination to DRIP handling and SG dynamics, which may stimulate new research directions and collaborative efforts across complementary areas of cell biology and molecular biology.
- Please define your field of expertise with a few keywords to help the authors contextualize your point of view. Indicate if there are any parts of the paper that you do not have sufficient expertise to evaluate.
I work in ubiquitin biology, focusing on ubiquitination signaling in physiological and disease contexts, with particular expertise in the identification of E3 ligases and their substrates across different cellular systems and in vivo models. I have less expertise in stress granule dynamics and DRiP biology, so my evaluation of those aspects is more limited and relies on interpretation of the data presented in the manuscript.
Reviewer #2 (Evidence, reproducibility and clarity (Required)):
This study identifies the E3 ubiquitin ligase Makorin 2 (MKRN2) as a novel regulator of stress granule (SG) dynamics and proteostasis. Using APEX proximity proteomics, the authors demonstrate that inhibition of the ubiquitin-activating enzyme UBA1 with TAK243 alters the SG proteome, leading to depletion of several E3 ligases, chaperones, and VCP cofactors. Detailed characterization of MKRN2 reveals that it localizes to SGs in a ubiquitination-dependent manner and is required for proper SG assembly, coalescence, and disassembly. Functionally, MKRN2 prevents the accumulation of defective ribosomal products (DRiPs) within SGs, thereby maintaining granulostasis. The study provides compelling evidence that ubiquitination, mediated specifically by MKRN2, plays a critical role in surveilling stress-damaged proteins within SGs and maintaining their dynamic liquid-like properties. Major issues: 1. Figures 1-2: Temporal dynamics of ubiquitination in SGs. The APEX proteomics was performed at a single timepoint (90 min heat stress), yet the live imaging data show that SG dynamics and TAK243 effects vary considerably over time: • The peak or SG nucleation was actually at 10-30 min (Figure 1B). • TAK243 treatment causes earlier SG nucleation (Figure 1B) but delayed disassembly (Figure 1A-B, D). A temporal proteomic analysis at multiple timepoints (e.g., 30 min, 60 min, 90 min of heat stress, and during recovery) would reveal whether MKRN2 and other ubiquitination-dependent proteins are recruited to SGs dynamically during the stress response. It would also delineate whether different E3 ligases predominate at different stages of the SG lifecycle. While such experiments may be beyond the scope of the current study, the authors should at minimum discuss this limitation and acknowledge that the single-timepoint analysis may miss dynamic changes in SG composition. *
Authors: We thank the reviewer for identifying this caveat in our methodology. We now discuss this limitation and acknowledge that the single-timepoint analysis may miss dynamic changes in SG composition.
* Figures 2D-E, 3G: MKRN2 localization mechanism requires clarification. The authors demonstrate that MKRN2 localization to SGs is dependent on active ubiquitination, as TAK243 treatment significantly reduces MKRN2 partitioning into SGs (Figure 2D-E). However, several mechanistic questions remain: • Does MKRN2 localize to SGs through binding to ubiquitinated substrates within SGs, or does MKRN2 require its own ubiquitination activity to enter SGs? • The observation that MKRN2 overexpression increases SG ubiquitin content (Figure 3G-H) could indicate either: (a) MKRN2 actively ubiquitinates substrates within SGs, or (b) MKRN2 recruitment brings along pre-ubiquitinated substrates from the cytoplasm. • Is MKRN2 localization to SGs dependent on its E3 ligase activity? A catalytically inactive mutant of MKRN2 would help distinguish whether MKRN2 must actively ubiquitinate proteins to remain in SGs or whether it binds to ubiquitinated proteins independently of its catalytic activity. The authors should clarify whether MKRN2's SG localization depends on its catalytic activity or on binding to ubiquitinated proteins, as this would fundamentally affect the interpretation of its role in SG dynamics. *
Authors: We thank the reviewer for this experimental suggestion. We will perform an analysis of the SG partitioning coefficient between WT-MKRN2 and a RING mutant of MKRN2.
* Figures 3-4: Discrepancy between assembly and disassembly phenotypes. MKRN2 knockdown produces distinct phenotypes during SG assembly versus disassembly. During assembly: smaller, more numerous SGs that fail to coalesce (Figure 3A-E), while during disassembly: delayed SG clearance (Figure 4A-D). These phenotypes may reflect different roles for MKRN2 at different stages, but the mechanism underlying this stage-specificity is unclear: • Does MKRN2 have different substrates or utilize different ubiquitin chain types during assembly versus disassembly? • The increased SG liquidity upon MKRN2 depletion (Figure 3F) seems paradoxical with delayed disassembly- typically more liquid condensates disassemble faster. The authors interpret this as decreased coalescence into "dense and mature SGs," but this requires clarification. • How does prevention of DRiP accumulation relate to the assembly defect? One would predict that DRiP accumulation would primarily affect disassembly (by reducing liquidity), yet MKRN2 depletion impacts both assembly dynamics and DRiP accumulation. The authors should discuss how MKRN2's role in preventing DRiP accumulation mechanistically connects to both the assembly and disassembly phenotypes. *
Authors: We thank the reviewer and will add to the Discussion a mention of a precedent for this precise phenotype from our previous work (Seguin et al., 2014).
* Figure 5: Incomplete characterization of MKRN2 substrates. While the authors convincingly demonstrate that MKRN2 prevents DRiP accumulation in SGs (Figure 5C-D), the direct substrates of MKRN2 remain unknown. The authors acknowledge in the limitations that "the direct MKRN2 substrates and ubiquitin-chain types (K63/K48) are currently unknown." However, several approaches could strengthen the mechanistic understanding: • Do DRiPs represent direct MKRN2 substrates? Co-immunoprecipitation of MKRN2 followed by ubiquitin-chain specific antibodies (K48 vs K63) could reveal whether MKRN2 mediates degradative (K48) or non-degradative (K63) ubiquitination. *
Authors: The DRiPs generated in the study represent truncated versions of all the proteins that were in the process of being synthesized by the cell at the moment of the stress, and therefore include both MKRN2 specific substrates and MKRN2 independent substrates. Identifying specific MKRN2 substrates, while interesting as a new research avenue, is not within the scope of the present study.
-
* Given that VCP cofactors (such as UFD1L, PLAA) are depleted from SGs upon UBA1 inhibition (Figure 2C) and these cofactors recognize ubiquitinated substrates, does MKRN2 function upstream of VCP recruitment? Testing whether MKRN2 depletion affects VCP cofactor localization to SGs would clarify this pathway. * Authors: We thank the reviewer for requesting and will address it by performing MKRN2 KD, VCP immunofluorescence microscopy and perform SG partition coefficient analysis.
-
* The authors note that MKRN2 knockdown produces a phenotype reminiscent of VCP inhibition-smaller, more numerous SGs with increased DRiP partitioning. This similarity suggests MKRN2 may function in the same pathway as VCP. Direct epistasis experiments would strengthen this connection. * Authors: This study is conditional results of the above study. If VCP partitioning to SGs is reduced upon MKRN2 KD, which we do not know at this point, then MKRN2/VCP double KD experiment will be performed to strengthen this connection.
* Alternative explanations for the phenotype of delayed disassembly with TAK243 or MKRN2 depletion- the authors attribute this to DRiP accumulation, but TAK243 affects global ubiquitination. Could impaired degradation of other SG proteins (not just DRiPs) contribute to delayed disassembly? Does proteasome inhibition (MG-132 treatment) phenocopy the MKRN2 depletion phenotype? This would support that MKRN2-mediated proteasomal degradation (via K48 ubiquitin chains) is key to the phenotype. *
Authors: We are happy to provide alternative explanations in the Discussion in line with Reviewer #2 suggestion. The role of the proteosome is out of the scope of our study.
-
Comparison with other E3 ligases (Supplementary Figure 5): The authors show that CNOT4 and ZNF598 depletion also affect SG dynamics, though to lesser extents than MKRN2. However: • Do these E3 ligases also prevent DRiP accumulation in SGs? Testing OP-puro partitioning in CNOT4- or ZNF598-depleted cells would reveal whether DRiP clearance is a general feature of SG-localized E3 ligases or specific to MKRN2. *
-
* Are there redundant or compensatory relationships between these E3 ligases? Do double knockdowns have additive effects? * Authors: Our paper presents a study of the E3 ligase MKRN2. Generalizing these observations to ZNF598, CNOT4 and perhaps an even longer list of E3s, may be an interesting question, outside the scope of our mission.
-
* The authors note that MKRN2 is "the most highly SG-depleted E3 upon TAK243 treatment"-does this mean MKRN2 has the strongest dependence on active ubiquitination for its SG localization, or simply that it has the highest basal level of SG partitioning? * Authors: We thank the reviewer for this smart question. MKRN2 has the strongest dependence on active ubiquitination as we now clarify better in the Results.
*Reviewer #2 (Significance (Required)):
This is a well-executed study that identifies MKRN2 as an important regulator of stress granule dynamics and proteostasis. The combination of proximity proteomics, live imaging, and functional assays provides strong evidence for MKRN2's role in preventing DRiP accumulation and maintaining granulostasis. However, key mechanistic questions remain, particularly regarding MKRN2's direct substrates, the ubiquitin chain types it generates, and how its enzymatic activity specifically prevents DRiP accumulation while promoting both SG coalescence and disassembly. Addressing the suggested revisions, particularly those related to MKRN2's mechanism of SG localization and substrate specificity, would significantly strengthen the manuscript and provide clearer insights into how ubiquitination maintains the dynamic properties of stress granules under proteotoxic stress.
Reviewer #3 (Evidence, reproducibility and clarity (Required)):
In this paper, Amzallag et al. investigate the relationship between ubiquitination and the dynamics of stress granules (SGs). They utilize proximity ligation coupled mass spectrometry to identify SG components under conditions where the proteasome is inhibited by a small drug that targets UBiquitin-like modifier Activating enzyme 1 (UBA1), which is crucial for the initial step in the ubiquitination of misfolded proteins. Their findings reveal that the E3 ligase Makorin2 (MKRN2) is a novel component of SGs. Additionally, their data suggest that MKRN2 is necessary for processing damaged ribosome-associated proteins (DRIPs) during heat shock (HS). In the absence of MKRN2, DRIPs accumulate in SGs, which affects their dynamics. Major comments: Assess the knockdown efficiency (KD) for CNOT1, ZNF598, and MKRN2 to determine if the significant effect observed on SG dynamics upon MKRN2 depletion is due to the protein's function rather than any possible differences in KD efficiency. *
Authors: To address potential variability in knockdown efficiency, we will quantify CNOT4, ZNF598, and MKRN2 mRNA levels by RT-qPCR following siRNA knockdown.
* Since HS-induced stress granules (SGs) are influenced by the presence of TAK-243 or MKRN2 depletion, could it be that these granules become more mature and thus acquire more defective ribosomal products (DRIPs)? Do HS cells reach the same level of DRIPs, as assessed by OP-Puro staining, at a later time point? *
Authors: an interesting question. Mateju et al. carefully characterized the time course of DRiP accumulation in stress granules during heat shock, decreasing after the 90 minutes point (Appendix Figure S7; 10.15252/embj.201695957). We therefore interpret DRiP accumulation in stress granules following TAK243 treatment as a pathological state, reflecting impaired removal and degradation of DRiPs, rather than a normal, more “mature” stress granule state.
* Incorporating OP-Puro can lead to premature translation termination, potentially confounding results. Consider treating cells with a short pulse (i.e., 5 minutes) of OP-Puro just before fixation. *
Authors: Thank you for this suggestion. Treating the cell with a short pulse of OP-Puro just before fixation will lead to the labelling of a small amount of proteins, likely undetectable using conventional microscopy or Western blotting. Furthermore, it will lead to the unwanted labeling of stress responsive proteins that are translated with non canonical cap-independent mechanisms upon stress.
* Is MKRN2's dependence limited to HS-induced SGs? *
Authors: We will test sodium arsenite–induced stress and use immunofluorescence at discrete time points to assess whether the heat shock–related observations generalize to other stress types.
*
Minor comments: Abstract: Introduce UBA1. Introduction: The reference [2] should be replaced with 25719440. Results: Line 70, 'G3BP1 and 2 genes,' is somewhat misleading. Consider rephrasing into 'G3BP1 and G3BP2 genes'. Line 103: considers rephrasing 'we orthogonally validated the ubiquitin-dependent interaction' to 'we orthogonally validated the ubiquitin-dependent stress granule localization'. Line 125: '(fig.3C, EI Supplementary fig. 3)' Remove 'I'. Methods: line 260: the reference is not linked (it should be ref. [26]). Line 225: Are all the KDs being performed using the same method? Please specify. *
Authors: The text has been altered to reflect the reviewer’s suggestions.
*Fig.2C: Consider adding 'DEPLETED' on top of the scheme.
Reviewer #3 (Significance (Required)):
The study offers valuable insights into the degradative processes associated with SGs. The figures are clear, and the experimental quality is high. The authors do not overstate or overinterpret their findings, and the results effectively support their claims. However, the study lacks orthogonal methods to validate the findings and enhance the results. For instance, incorporating biochemical and reporter-based methods to measure degradation-related intermediate products (DRIPs) would be beneficial. Additionally, utilizing multiple methods to block ubiquitination, studying the dynamics of MKRN2 on SGs, and examining the consequences of excessive DRIPs on the cell fitness of SGs would further strengthen the research. *
-
Note: This preprint has been reviewed by subject experts for Review Commons. Content has not been altered except for formatting.
Learn more at Review Commons
Referee #1
Evidence, reproducibility and clarity
Summary:
In this study, the authors used proximity proteomics in U2OS cells to identify several E3 ubiquitin ligases recruited to stress granules (SGs), and they focused on MKRN2 as a novel regulator. They show that MKRN2 localization to SGs requires active ubiquitination via UBA1. Functional experiments demonstrated that MKRN2 knockdown increases the number of SG condensates, reduces their size, slightly raises SG liquidity during assembly, and slows disassembly after heat shock. Overexpression of MKRN2-GFP combined with confocal imaging revealed co-localization of MKRN2 and ubiquitin in SGs. By perturbing ubiquitination (using a UBA1 inhibitor) and inducing defective ribosomal products (DRiPs) with O-propargyl puromycin, they found that both ubiquitination inhibition and MKRN2 depletion lead to increased accumulation of DRiPs in SGs. The authors conclude that MKRN2 supports granulostasis, the maintenance of SG homeostasis , through its ubiquitin ligase activity, preventing pathological DRiP accumulation within SGs.
Major comments:
- Are the key conclusions convincing?
The key conclusions are partially convincing. The data supporting the role of ubiquitination and MKRN2 in regulating SG condensate dynamics are coherent, well controlled, and consistent with previous literature, making this part of the study solid and credible. However, the conclusions regarding the ubiquitin-dependent recruitment of MKRN2 to SGs, its relationship with UBA1 activity, the functional impact of the MKRN2 knockdown for DRiP accumulation are less thoroughly supported. These aspects would benefit from additional mechanistic evidence, validation in complementary model systems, or the use of alternative methodological approaches to strengthen the causal connections drawn by the authors. - Should the authors qualify some of their claims as preliminary or speculative, or remove them altogether? The authors should qualify some of their claims as preliminary.
1) MKRN2 recruitment to SGs (ubiquitin-dependent): The proteomics and IF data are a reasonable starting point, but they do not yet establish that MKRN2 is recruited from its physiological localization to SGs in a ubiquitin-dependent manner. To avoid overstating this point the authors should qualify the claim and/or provide additional controls: show baseline localization of endogenous MKRN2 under non-stress conditions (which is reported in literature to be nuclear and cytoplasmatic), include quantification of nuclear/cytoplasmic distribution, and demonstrate a shift into bona fide SG compartments after heat shock. Moreover, co-localization of overexpressed GFP-MKRN2 with poly-Ub (FK2) should be compared to a non-stress control and to UBA1-inhibition conditions to support claims of stress- and ubiquitination-dependent recruitment.
2) Use and interpretation of UBA1 inhibition: UBA1 inhibition effectively blocks ubiquitination globally, but it is non-selective. The manuscript should explicitly acknowledge this limitation when interpreting results from both proteomics and functional assays. Proteomics hits identified under UBA1 inhibition should be discussed as UBA1-dependent associations rather than as evidence for specific E3 ligase recruitment. The authors should consider orthogonal approaches before concluding specificity.
3) DRiP accumulation and imaging quality: The evidence presented in Figure 5 is sufficient to substantiate the claim that DRiPs accumulate in SGs upon ubiquitination inhibition or MKRN2 depletion but to show that the event of the SGs localization and their clearance from SGs during stress is promoted by MKRN3 ubiquitin ligase activity more experiments would be needed. - Would additional experiments be essential to support the claims of the paper? Request additional experiments only where necessary for the paper as it is, and do not ask authors to open new lines of experimentation. Yes, a few targeted experiments would strengthen the conclusions without requiring the authors to open new lines of investigation.
1) Baseline localization of MKRN2: It would be important to show the baseline localization of endogenous and over-expressed MKRN2 (nuclear and cytoplasmic) under non-stress conditions and prior to ubiquitination inhibition. This would provide a reference to quantify redistribution into SGs and demonstrate recruitment in response to heat stress or ubiquitination-dependent mechanisms.
2) Specificity of MKRN2 ubiquitin ligase activity: to address the non-specific effects of UBA1 inhibition and validate that observed phenotypes depend on MKRN2's ligase activity, the authors could employ a catalytically inactive MKRN2 mutant in rescue experiments. Comparing wild-type and catalytic-dead MKRN2 in the knockdown background would clarify the causal role of MKRN2 activity in SG dynamics and DRiP clearance.
3) Ubiquitination linkage and SG marker levels: While the specific ubiquitin linkage type remains unknown, examining whether MKRN2 knockdown or overexpression affects total levels of key SG marker proteins would be informative. This could be done via Western blotting of SG markers along with ubiquitin staining, to assess whether MKRN2 influences protein stability or turnover through degradative or non-degradative ubiquitination. Such data would strengthen the mechanistic interpretation while remaining within the current study's scope. - Are the suggested experiments realistic in terms of time and resources? It would help if you could add an estimated cost and time investment for substantial experiments. The experiments suggested in points 1 and 3 are realistic and should not require substantial additional resources beyond those already used in the study. - Point 1 (baseline localization of MKRN2): This involves adding two control conditions (no stress and no ubiquitination inhibition) for microscopy imaging. The setup is essentially the same as in the current experiments, with time requirements mainly dependent on cell culture growth and imaging. Overall, this could be completed within a few weeks. - Point 3 (SG marker levels and ubiquitination): This entails repeating the existing experiment and adding a Western blot for SG markers and ubiquitin. The lab should already have the necessary antibodies, and the experiment could reasonably be performed within a couple of weeks. - Point 2 (catalytically inactive MKRN2 mutant and rescue experiments): This is likely more time-consuming. Designing an effective catalytic-dead mutant depends on structural knowledge of MKRN2 and may require additional validation to confirm loss of catalytic activity. If this expertise is not already present in the lab, it could significantly extend the timeline. Therefore, this experiment should be considered only if similarly recommended by other reviewers, as it represents a higher resource and time investment.
Overall, points 1 and 3 are highly feasible, while point 2 is more substantial and may require careful planning. - Are the data and the methods presented in such a way that they can be reproduced?
Yes. The methodologies used in this study to analyze SG dynamics and DRiP accumulation are well-established in the field and should be reproducible, particularly by researchers experienced in stress granule biology. Techniques such as SG assembly and disassembly assays, use of G3BP1 markers, and UBA1 inhibition are standard and clearly described. The data are generally presented in a reproducible manner; however, as noted above, some results would benefit from additional controls or complementary experiments to fully support specific conclusions. - Are the experiments adequately replicated and statistical analysis adequate?
Overall, the experiments in the manuscript appear to be adequately replicated, with most assays repeated between three and five times, as indicated in the supplementary materials. The statistical analyses used are appropriate and correctly applied to the datasets presented. However, for Figure 5 the number of experimental replicates is not reported. This should be clarified, and if the experiment was not repeated sufficiently, additional biological replicates should be performed. Given that this figure provides central evidence supporting the conclusion that DRiP accumulation depends on ubiquitination-and partly on MKRN2's ubiquitin ligase activity-adequate replication is essential.
Minor comments:
- Specific experimental issues that are easily addressable.
- For the generation and the validation of MKRN2 knockdown in UOS2 cells data are not presented in the results or in the methods sections to demonstrate the effective knockdown of the protein of interest. This point is quite essential to demonstrate the validity of the system used
- In the supplementary figure 2 it would be useful to mention if the Western Blot represent the input (total cell lysates) before the APEX-pulldown or if it is the APEX-pulldown loaded for WB. There is no consistence in the difference of biotynilation between different replicates shown in the 2 blots. For example in R1 and R2 G3BP1-APX TAK243 the biotynilation is one if the strongest condition while on the left blot, in the same condition comparison samples R3 and R4 are less biotinilated compared to others. It would be useful to provide an explanation for that to avoid any confusion for the readers.
- In Figure 2D, endogenous MKRN2 localization to SGs appears reduced following UBA1 inhibition. However, it is not clear whether this reduction reflects a true relocalization or a decrease in total MKRN2 protein levels. To support the interpretation that UBA1 inhibition specifically affects MKRN2 recruitment to SGs rather than its overall expression, the authors should provide data showing total MKRN2 levels remain unchanged under UBA1 inhibition, for example via Western blot of total cell lysates.
- DRIPs accumulation is followed during assembly but in the introduction is highlighted the fact that ubiquitination events, other reported E3 ligases and in this study data on MKRN2 showed that they play a crucial role in the disassembly of SGs which is also related with cleareance of DRIPs. Authors could add tracking DRIPs accumulation during disassembly to be added to Figure 5. I am not sure about the timeline required for this but I am just adding as optional if could be addressed easily.
- The authors should clarify in the text why the cutoff used for the quantification in Figure 5D (PC > 3) differs from the cutoff used elsewhere in the paper (PC > 1.5). Providing a rationale for this choice will help the reader understand the methodological consistency and ensure that differences in thresholds do not confound interpretation of the results.
- For Figure 3G, the authors use over-expressed MKRN2-GFP to assess co-localization with ubiquitin in SGs. Given that a reliable antibody for endogenous MKRN2 is available and that a validated MKRN2 knockdown line exists as an appropriate control, this experiment would gain significantly in robustness and interpretability if co-localization were demonstrated using endogenous MKRN2. In the current over-expression system, MKRN2-GFP is also present in the nucleus, whereas the endogenous protein does not appear nuclear under the conditions shown. This discrepancy raises concerns about potential over-expression artifacts or mislocalization. Demonstrating co-localization using endogenous MKRN2 would avoid confounding effects associated with over-expression. If feasible, this would be a relatively straightforward experiment to implement, as it relies on tools (antibody and knockdown line) already described in the manuscript.
-
Are prior studies referenced appropriately?
- From line 54 to line 67, the manuscript in total cites eight papers regarding the role of ubiquitination in SG disassembly. However, given the use of UBA1 inhibition in the initial MS-APEX experiment and the extensive prior literature on ubiquitination in SG assembly and disassembly under various stress conditions, the manuscript would benefit from citing additional relevant studies to provide more specifc examples. Expanding the references would provide stronger context, better connect the current findings to prior work, and emphasize the significance of the study in relation to established literature
- At line 59, it would be helpful to note that G3BP1 is ubiquitinated by TRIM21 through a Lys63-linked ubiquitin chain. This information provides important mechanistic context, suggesting that ubiquitination of SG proteins in these pathways is likely non-degradative and related to functional regulation of SG dynamics rather than protein turnover.
- When citing references 16 and 17, which report that the E3 ligases TRIM21 and HECT regulate SG formation, the authors should provide a plausible explanation for why these specific E3 ligases were not detected in their proteomics experiments. Differences could arise from the stress stimulus used, cell type, or experimental conditions. Similarly, since MKRN2 and other E3 ligases identified in this study have not been reported in previous works, discussing these methodological or biological differences would help prevent readers from questioning the credibility of the findings. It would also be valuable to clarify in the Conclusion that different types of stress may activate distinct ubiquitination pathways, highlighting context-dependent regulation of SG assembly and disassembly.
- Line 59-60: when referring to the HECT family of E3 ligases involved in ubiquitination and SG disassembly, it would be more precise to report the specific E3 ligase identified in the cited studies rather than only the class of ligase. This would provide clearer mechanistic context and improve accuracy for readers.
- The specific statement on line 182 "SG E3 ligases that depend on UBA1 activity are RBULs" should be supported by reference.
- Are the text and figures clear and accurate?
- In Supplementary Figure 1, DMSO is shown in green and the treatment in red, whereas in the main figures (Figure 1B and 1F) the colours in the legend are inverted. To avoid confusion, the colour coding in figure legends should be consistent across all figures throughout the manuscript.
- At line 79, the manuscript states that "inhibition of ubiquitination delayed fluorescence recovery dynamics of G3BP1-mCherry, relative to HS-treated cells (Figure 1F, Supplementary Fig. 6A)." However, the data shown in Figure 1F appear to indicate the opposite effect: the TAK243-treated condition (green curve) shows a faster fluorescence recovery compared to the control (red curve). This discrepancy between the text and the figure should be corrected or clarified, as it may affect the interpretation of the role of ubiquitination in SG dynamics.
- Line 86: adjust a missing bracket
- There appears to be an error in the legend of Supplementary Figure 3: the legend states that the red condition (MKRN2) forms larger aggregates, but both the main Figure 3C of the confocal images and the text indicate that MKRN2 (red) forms smaller aggregates. Please correct the legend and any corresponding labels so they are consistent with the main figure and the text. The authors should also double-check that the figure panel order, color coding, and statistical annotations match the legend and the descriptions in the Results section to avoid reader confusion.
- At lines 129-130, the manuscript states that "FRAP analysis demonstrated that MKRN2 KD resulted in a slight increase in SG liquidity (Fig. 3F, Supplementary Fig. 6B)." However, the data shown in Figure 3F appear to indicate the opposite trend: the MKRN2 KD condition (red curve) exhibits a faster fluorescence recovery compared to the control (green curve). This discrepancy between the text and the figure should be corrected or clarified, as it directly affects the interpretation of MKRN2's role in SG disassembly. Ensuring consistency between the written description and the plotted FRAP data is essential for accurate interpretation.
- At lines 132-133, the manuscript states: "Then, to further test the impact of MKRN2 on SG dynamics, we overexpressed MKRN2-GFP and observed that it was recruited to SG (Fig. 3G)." This description should be corrected or clarified, as the over-expressed MKRN2-GFP also appears to localize to the nucleus.
- At lines 134-135, the manuscript states that the FK2 antibody detects "free ubiquitin." This is incorrect. FK2 does not detect free ubiquitin; it recognizes only ubiquitin conjugates, including mono-ubiquitinated and poly-ubiquitinated proteins. The text should be corrected accordingly to avoid misinterpretation of the immunostaining data.
- Figure 5A suffers from poor resolution, and no scale bar is provided, which limits interpretability. Additionally, the ROI selected for the green channel (DRIPs) appears to capture unspecific background staining, while the most obvious DRIP spots are localized in the nucleus. The authors should clarify this in the text, improve the image quality if possible, and ensure that the ROI accurately represents DRIP accumulation - in SGs rather than background signal.
Do you have suggestions that would help the authors improve the presentation of their data and conclusions?
- In the first paragraph following the APEX proteomics results, the authors present validation data exclusively for MKRN2, justifying this early focus by stating that MKRN2 is the most SG-depleted E3 ligase. However, in the subsequent paragraph they introduce the RBULs and present knockdown data for MKRN2 along with two additional E3 ligases identified in the screen, before once again emphasizing that MKRN2 is the most SG-depleted ligase and therefore the main focus of the study. For clarity and logical flow, the manuscript would benefit from reordering the narrative. Specifically, the authors should first present the validation data for all three selected E3 ligases, and only then justify the decision to focus on MKRN2 for in-depth characterization. In addition to the extent of its SG depletion, the authors may also consider providing biologically relevant reasons for prioritizing MKRN2 (e.g., domain architecture, known roles in stress responses, or prior evidence of ubiquitination-related functions). Reorganizing this section would improve readability and better guide the reader through the rationale for the study's focus.
- At lines 137-138, the manuscript states: "Together these data indicate that MKRN2 regulates the assembly dynamics of SGs by promoting their coalescence during HS and can increase SG ubiquitin content." While Figure 3G shows some co-localization of MKRN2 with ubiquitin, immunofluorescence alone is insufficient to claim an increase in SG ubiquitin content. This conclusion should be supported by orthogonal experiments, such as Western blotting, in vitro ubiquitination assays, or immunoprecipitation of SG components. Including a control under no-stress conditions would also help demonstrate that ubiquitination increases specifically in response to stress. The second part of the statement should therefore be rephrased to avoid overinterpretation, for example:"...and may be associated with increased ubiquitination within SGs, as suggested by co-localization, pending further validation by complementary assays."
- At line 157, the statement: "Therefore, we conclude that MKRN2 ubiquitinates a subset of DRiPs, avoiding their accumulation inside SGs" should be rephrased as a preliminary observation. While the data support a role for MKRN2 in SG disassembly and a reduction of DRIPs, direct ubiquitination of DRIPs by MKRN2 has not been demonstrated. A more cautious phrasing would better reflect the current evidence and avoid overinterpretation.
Significance
General assessment: provide a summary of the strengths and limitations of the study. What are the strongest and most important aspects? What aspects of the study should be improved or could be developed?
- This study provides a valuable advancement in understanding the role of ubiquitination in stress granule (SG) dynamics and the clearance of SGs formed under heat stress. A major strength is the demonstration of how E3 ligases identified through proteomic screening, particularly MKRN2, influence SG assembly and disassembly in a ubiquitination- and heat stress-dependent manner. The combination of proteomics, imaging, and functional assays provides a coherent mechanistic framework linking ubiquitination to SG homeostasis. Limitations of the study include the exclusive use of a single model system (U2OS cells), which may limit generalizability. Additionally, some observations-such as MKRN2-dependent ubiquitination within SGs and changes in DRIP accumulation under different conditions-would benefit from orthogonal validation experiments (e.g., Western blotting, immunoprecipitation, or in vitro assays) to confirm and strengthen these findings. Addressing these points would enhance the robustness and broader applicability of the conclusions.
Advance: compare the study to the closest related results in the literature or highlight results reported for the first time to your knowledge; does the study extend the knowledge in the field and in which way? Describe the nature of the advance and the resulting insights (for example: conceptual, technical, clinical, mechanistic, functional,...).
- The closest related result in literature is - Yang, Cuiwei et al. "Stress granule homeostasis is modulated by TRIM21-mediated ubiquitination of G3BP1 and autophagy-dependent elimination of stress granules." Autophagy vol. 19,7 (2023): 1934-1951. doi:10.1080/15548627.2022.2164427 - demonstrating that TRIM21, an E3 ubiquitin ligase, catalyzes K63-linked ubiquitination of G3BP1, a core SG nucleator, under oxidative stress. This ubiquitination by TRIM21 inhibits SG formation, likely by altering G3BP1's propensity for phase separation. In contrast, the MKRN2 study identifies a different E3 (MKRN2) that regulates SG dynamics under heat stress and appears to influence both assembly and disassembly. This expands the role of ubiquitin ligases in SG regulation beyond those previously studied (like TRIM21).
- Gwon and colleagues (Gwon Y, Maxwell BA, Kolaitis RM, Zhang P, Kim HJ, Taylor JP. Ubiquitination of G3BP1 mediates stress granule disassembly in a context-specific manner. Science. 2021;372(6549):eabf6548. doi:10.1126/science.abf6548) have shown that K63-linked ubiquitination of G3BP1 is required for SG disassembly after heat stress. This ubiquitinated G3BP1 recruits the segregase VCP/p97, which helps extract G3BP1 from SGs for disassembly. The MKRN2 paper builds on this by linking UBA1-dependent ubiquitination and MKRN2's activity to SG disassembly. Specifically, they show MKRN2 knockdown affects disassembly, and suggest MKRN2 helps prevent accumulation of defective ribosomal products (DRiPs) in SGs, adding a new layer to the ubiquitin-VCP model.
- Ubiquitination's impact is highly stress- and context-dependent (different chain types, ubiquitin linkages, and recruitment of E3s). The MKRN2 work conceptually strengthens this idea: by showing that MKRN2's engagement with SGs depends on active ubiquitination via UBA1, and by demonstrating functional consequences (SG dynamics + DRIP accumulation), the study highlights how cellular context (e.g., heat stress) can recruit specific ubiquitin ligases to SGs and modulate their behavior.
- There is a gap in the literature: very few (if any) studies explicitly combine the biology of DRIPs, stress granules, and E3 ligase mediated ubiquitination, especially in mammalian cells. There are relevant works about DRIP biology in stress granules, but those studies focus on chaperone-based quality control, not ubiquitin ligase-mediated ubiquitination of DRIPs. This study seems to be one of the first to make that connection in mammalian (or human-like) SG biology. A work on the plant DRIP-E3 ligase TaSAP5 (Zhang N, Yin Y, Liu X, et al. The E3 Ligase TaSAP5 Alters Drought Stress Responses by Promoting the Degradation of DRIP Proteins. Plant Physiol. 2017;175(4):1878-1892. doi:10.1104/pp.17.01319 ) shows that DRIPs can be directly ubiquitinated by E3s in other biological systems - which supports the plausibility of the MKRN2 mechanism, but it's not the same context.
- A very recent review (Yuan, Lin et al. "Stress granules: emerging players in neurodegenerative diseases." Translational neurodegeneration vol. 14,1 22. 12 May. 2025, doi:10.1186/s40035-025-00482-9) summarizes and reinforces the relationship among SGs and the pathogenesis of different neurodegenerative diseases (NDDs). By identifying MKRN2 as a new ubiquitin regulator in SGs, the current study could have relevance for neurodegeneration and proteotoxic diseases, providing a new candidate to explore in disease models.
Audience: describe the type of audience ("specialized", "broad", "basic research", "translational/clinical", etc...) that will be interested or influenced by this research; how will this research be used by others; will it be of interest beyond the specific field?
The audience for this paper is primarily specialized, including researchers in stress granule biology, ubiquitin signaling, protein quality control, ribosome biology, and cellular stress responses. The findings will also be of interest to scientists working on granulostasis, nascent protein surveillance, and proteostasis mechanisms. Beyond these specific fields, the study provides preliminary evidence linking ubiquitination to DRIP handling and SG dynamics, which may stimulate new research directions and collaborative efforts across complementary areas of cell biology and molecular biology.
Please define your field of expertise with a few keywords to help the authors contextualize your point of view. Indicate if there are any parts of the paper that you do not have sufficient expertise to evaluate.
I work in ubiquitin biology, focusing on ubiquitination signaling in physiological and disease contexts, with particular expertise in the identification of E3 ligases and their substrates across different cellular systems and in vivo models. I have less expertise in stress granule dynamics and DRiP biology, so my evaluation of those aspects is more limited and relies on interpretation of the data presented in the manuscript.
-
-
www.biorxiv.org www.biorxiv.org
-
Reviewer #1 (Public review):
Summary:
Zhou and colleagues developed a computational model of replay that heavily builds on cognitive models of memory in context (e.g., the context-maintenance and retrieval model), which have been successfully used to explain memory phenomena in the past. Their model produces results that mirror previous empirical findings in rodents and offers a new computational framework for thinking about replay.
Strengths:
The model is compelling and seems to explain a number of findings from the rodent literature. It is commendable that the authors implement commonly used algorithms from wakefulness to model sleep/rest, thereby linking wake and sleep phenomena in a parsimonious way. Additionally, the manuscript's comprehensive perspective on replay, bridging humans and non-human animals, enhanced its theoretical contribution.
Weaknesses:
This reviewer is not a computational neuroscientist by training, so some comments may stem from misunderstandings. I hope the authors would see those instances as opportunities to clarify their findings for broader audiences.
(1) The model predicts that temporally close items will be co-reactivated, yet evidence from humans suggests that temporal context doesn't guide sleep benefits (instead, semantic connections seem to be of more importance; Liu and Ranganath 2021, Schechtman et al 2023). Could these findings be reconciled with the model or is this a limitation of the current framework?
(2) During replay, the model is set so that the next reactivated item is sampled without replacement (i.e., the model cannot get "stuck" on a single item). I'm not sure what the biological backing behind this is and why the brain can't reactivate the same item consistently. Furthermore, I'm afraid that such a rule may artificially generate sequential reactivation of items regardless of wake training. Could the authors explain this better or show that this isn't the case?
(3) If I understand correctly, there are two ways in which novelty (i.e., less exposure) is accounted for in the model. The first and more talked about is the suppression mechanism (lines 639-646). The second is a change in learning rates (lines 593-595). It's unclear to me why both procedures are needed, how they differ, and whether these are two different mechanisms that the model implements. Also, since the authors controlled the extent to which each item was experienced during wakefulness, it's not entirely clear to me which of the simulations manipulated novelty on an individual item level, as described in lines 593-595 (if any).
As to the first mechanism - experience-based suppression - I find it challenging to think of a biological mechanism that would achieve this and is selectively activated immediately before sleep (somehow anticipating its onset). In fact, the prominent synaptic homeostasis hypothesis suggests that such suppression, at least on a synaptic level, is exactly what sleep itself does (i.e., prune or weaken synapses that were enhanced due to learning during the day). This begs the question of whether certain sleep stages (or ultradian cycles) may be involved in pruning, whereas others leverage its results for reactivation (e.g., a sequential hypothesis; Rasch & Born, 2013). That could be a compelling synthesis of this literature. Regardless of whether the authors agree, I believe that this point is a major caveat to the current model. It is addressed in the discussion, but perhaps it would be beneficial to explicitly state to what extent the results rely on the assumption of a pre-sleep suppression mechanism.
(4) As the manuscript mentions, the only difference between sleep and wake in the model is the initial conditions (a0). This is an obvious simplification, especially given the last author's recent models discussing the very different roles of REM vs NREM. Could the authors suggest how different sleep stages may relate to the model or how it could be developed to interact with other successful models such as the ones the last author has developed (e.g., C-HORSE)? Finally, I wonder how the model would explain findings (including the authors') showing a preference for reactivation of weaker memories. The literature seems to suggest that it isn't just a matter of novelty or exposure, but encoding strength. Can the model explain this? Or would it require additional assumptions or some mechanism for selective endogenous reactivation during sleep and rest?
(5) Lines 186-200 - Perhaps I'm misunderstanding, but wouldn't it be trivial that an external cue at the end-item of Figure 7a would result in backward replay, simply because there is no potential for forward replay for sequences starting at the last item (there simply aren't any subsequent items)? The opposite is true, of course, for the first-item replay, which can't go backward. More generally, my understanding of the literature on forward vs backward replay is that neither is linked to the rodent's location. Both commonly happen at a resting station that is further away from the track. It seems as though the model's result may not hold if replay occurs away from the track (i.e. if a0 would be equal for both pre- and post-run).
(6) The manuscript describes a study by Bendor & Wilson (2012) and tightly mimics their results. However, notably, that study did not find triggered replay immediately following sound presentation, but rather a general bias toward reactivation of the cued sequence over longer stretches of time. In other words, it seems that the model's results don't fully mirror the empirical results. One idea that came to mind is that perhaps it is the R/L context - not the first R/L item - that is cued in this study. This is in line with other TMR studies showing what may be seen as contextual reactivation. If the authors think that such a simulation may better mirror the empirical results, I encourage them to try. If not, however, this limitation should be discussed.
(7) There is some discussion about replay's benefit to memory. One point of interest could be whether this benefit changes between wake and sleep. Relatedly, it would be interesting to see whether the proportion of forward replay, backward replay, or both correlated with memory benefits. I encourage the authors to extend the section on the function of replay and explore these questions.
(8) Replay has been mostly studied in rodents, with few exceptions, whereas CMR and similar models have mostly been used in humans. Although replay is considered a good model of episodic memory, it is still limited due to limited findings of sequential replay in humans and its reliance on very structured and inherently autocorrelated items (i.e., place fields). I'm wondering if the authors could speak to the implications of those limitations on the generalizability of their model. Relatedly, I wonder if the model could or does lead to generalization to some extent in a way that would align with the complementary learning systems framework.
-
Author response:
The following is the authors’ response to the original reviews.
Public Reviews:
Reviewer #1 (Public Review):
Summary:
Zhou and colleagues developed a computational model of replay that heavily builds on cognitive models of memory in context (e.g., the context-maintenance and retrieval model), which have been successfully used to explain memory phenomena in the past. Their model produces results that mirror previous empirical findings in rodents and offers a new computational framework for thinking about replay.
Strengths:
The model is compelling and seems to explain a number of findings from the rodent literature. It is commendable that the authors implement commonly used algorithms from wakefulness to model sleep/rest, thereby linking wake and sleep phenomena in a parsimonious way. Additionally, the manuscript's comprehensive perspective on replay, bridging humans and non-human animals, enhanced its theoretical contribution.
Weaknesses:
This reviewer is not a computational neuroscientist by training, so some comments may stem from misunderstandings. I hope the authors would see those instances as opportunities to clarify their findings for broader audiences.
(1) The model predicts that temporally close items will be co-reactivated, yet evidence from humans suggests that temporal context doesn't guide sleep benefits (instead, semantic connections seem to be of more importance; Liu and Ranganath 2021, Schechtman et al 2023). Could these findings be reconciled with the model or is this a limitation of the current framework?
We appreciate the encouragement to discuss this connection. Our framework can accommodate semantic associations as determinants of sleep-dependent consolidation, which can in principle outweigh temporal associations. Indeed, prior models in this lineage have extensively simulated how semantic associations support encoding and retrieval alongside temporal associations. It would therefore be straightforward to extend our model to simulate how semantic associations guide sleep benefits, and to compare their contribution against that conferred by temporal associations across different experimental paradigms. In the revised manuscript, we have added a discussion of how our framework may simulate the role of semantic associations in sleep-dependent consolidation.
“Several recent studies have argued for dominance of semantic associations over temporal associations in the process of human sleep-dependent consolidation (Schechtman et al., 2023; Liu and Ranganath 2021; Sherman et al., 2025), with one study observing no role at all for temporal associations (Schechtman et al., 2023). At first glance, these findings appear in tension with our model, where temporal associations drive offline consolidation. Indeed, prior models have accounted for these findings by suppressing temporal context during sleep (Liu and Ranganath 2024; Sherman et al., 2025). However, earlier models in the CMR lineage have successfully captured the joint contributions of semantic and temporal associations to encoding and retrieval (Polyn et al., 2009), and these processes could extend naturally to offline replay. In a paradigm where semantic associations are especially salient during awake learning, the model could weight these associations more and account for greater co-reactivation and sleep-dependent memory benefits for semantically related than temporally related items. Consistent with this idea, Schechtman et al. (2023) speculated that their null temporal effects likely reflected the task’s emphasis on semantic associations. When temporal associations are more salient and task-relevant, sleep-related benefits for temporally contiguous items are more likely to emerge (e.g., Drosopoulos et al., 2007; King et al., 2017).”
The reviewer’s comment points to fruitful directions for future work that could employ our framework to dissect the relative contributions of semantic and temporal associations to memory consolidation.
(2) During replay, the model is set so that the next reactivated item is sampled without replacement (i.e., the model cannot get "stuck" on a single item). I'm not sure what the biological backing behind this is and why the brain can't reactivate the same item consistently.
Furthermore, I'm afraid that such a rule may artificially generate sequential reactivation of items regardless of wake training. Could the authors explain this better or show that this isn't the case?
We appreciate the opportunity to clarify this aspect of the model. We first note that this mechanism has long been a fundamental component of this class of models (Howard & Kahana 2002). Many classic memory models (Brown et al., 2000; Burgess & Hitch, 1991; Lewandowsky & Murdock 1989) incorporate response suppression, in which activated items are temporarily inhibited. The simplest implementation, which we use here, removes activated items from the pool of candidate items. Alternative implementations achieve this through transient inhibition, often conceptualized as neuronal fatigue (Burgess & Hitch, 1991; Grossberg 1978). Our model adopts a similar perspective, interpreting this mechanism as mimicking a brief refractory period that renders reactivated neurons unlikely to fire again within a short physiological event such as a sharp-wave ripple. Importantly, this approach does not generate spurious sequences. Instead, the model’s ability to preserve the structure of wake experience during replay depends entirely on the learned associations between items (without these associations, item order would be random). Similar assumptions are also common in models of replay. For example, reinforcement learning models of replay incorporate mechanisms such as inhibition to prevent repeated reactivations (e.g., Diekmann & Cheng, 2023) or prioritize reactivation based on ranking to limit items to a single replay (e.g., Mattar & Daw, 2018). We now discuss these points in the section titled “A context model of memory replay”
“This mechanism of sampling without replacement, akin to response suppression in established context memory models (Howard & Kahana 2002), could be implemented by neuronal fatigue or refractory dynamics (Burgess & Hitch, 1991; Grossberg 1978). Non-repetition during reactivation is also a common assumption in replay models that regulate reactivation through inhibition or prioritization (Diekmann & Cheng 2023; Mattar & Daw 2018; Singh et al., 2022).”
(3) If I understand correctly, there are two ways in which novelty (i.e., less exposure) is accounted for in the model. The first and more talked about is the suppression mechanism (lines 639-646). The second is a change in learning rates (lines 593-595). It's unclear to me why both procedures are needed, how they differ, and whether these are two different mechanisms that the model implements. Also, since the authors controlled the extent to which each item was experienced during wakefulness, it's not entirely clear to me which of the simulations manipulated novelty on an individual item level, as described in lines 593-595 (if any).
We agree that these mechanisms and their relationships would benefit from clarification. As noted, novelty influences learning through two distinct mechanisms. First, the suppression mechanism is essential for capturing the inverse relationship between the amount of wake experience and the frequency of replay, as observed in several studies. This mechanism ensures that items with high wake activity are less likely to dominate replay. Second, the decrease in learning rates with repetition is crucial for preserving the stochasticity of replay. Without this mechanism, the model would increase weights linearly, leading to an exponential increase in the probability of successive wake items being reactivated back-to-back due to the use of a softmax choice rule. This would result in deterministic replay patterns, which are inconsistent with experimental observations.
We have revised the Methods section to explicitly distinguish these two mechanisms:
“This experience-dependent suppression mechanism is distinct from the reduction of learning rates through repetition; it does not modulate the update of memory associations but exclusively governs which items are most likely to initiate replay.”
We have also clarified our rationale for including a learning rate reduction mechanism:
“The reduction in learning rates with repetition is important for maintaining a degree of stochasticity in the model’s replay during task repetition, since linearly increasing weights would, through the softmax choice rule, exponentially amplify differences in item reactivation probabilities, sharply reducing variability in replay.”
Finally, we now specify exactly where the learning-rate reduction applied, namely in simulations where sequences are repeated across multiple sessions:
“In this simulation, the learning rates progressively decrease across sessions, as described above.“
As to the first mechanism - experience-based suppression - I find it challenging to think of a biological mechanism that would achieve this and is selectively activated immediately before sleep (somehow anticipating its onset). In fact, the prominent synaptic homeostasis hypothesis suggests that such suppression, at least on a synaptic level, is exactly what sleep itself does (i.e., prune or weaken synapses that were enhanced due to learning during the day). This begs the question of whether certain sleep stages (or ultradian cycles) may be involved in pruning, whereas others leverage its results for reactivation (e.g., a sequential hypothesis; Rasch & Born, 2013). That could be a compelling synthesis of this literature. Regardless of whether the authors agree, I believe that this point is a major caveat to the current model. It is addressed in the discussion, but perhaps it would be beneficial to explicitly state to what extent the results rely on the assumption of a pre-sleep suppression mechanism.
We appreciate the reviewer raising this important point. Unlike the mechanism proposed by the synaptic homeostasis hypothesis, the suppression mechanism in our model does not suppress items based on synapse strength, nor does it modify synaptic weights. Instead, it determines the level of suppression for each item based on activity during awake experience. The brain could implement such a mechanism by tagging each item according to its activity level during wakefulness. During subsequent consolidation, the initial reactivation of an item during replay would reflect this tag, influencing how easily it can be reactivated.
A related hypothesis has been proposed in recent work, suggesting that replay avoids recently active trajectories due to spike frequency adaptation in neurons (Mallory et al., 2024). Similarly, the suppression mechanism in our model is critical for explaining the observed negative relationship between the amount of recent wake experience and the degree of replay.
We discuss the biological plausibility of this mechanism and its relationship with existing models in the Introduction. In the section titled “The influence of experience”, we have added the following:
“Our model implements an activity‑dependent suppression mechanism that, at the onset of each offline replay event, assigns each item a selection probability inversely proportional to its activation during preceding wakefulness. The brain could implement this by tagging each memory trace in proportion to its recent activation; during consolidation, that tag would then regulate starting replay probability, making highly active items less likely to be reactivated. A recent paper found that replay avoids recently traversed trajectories through awake spike‑frequency adaptation (Mallory et al., 2025), which could implement this kind of mechanism. In our simulations, this suppression is essential for capturing the inverse relationship between replay frequency and prior experience. Note that, unlike the synaptic homeostasis hypothesis (Tononi & Cirelli 2006), which proposes that the brain globally downscales synaptic weights during sleep, this mechanism leaves synaptic weights unchanged and instead biases the selection process during replay.”
(4) As the manuscript mentions, the only difference between sleep and wake in the model is the initial conditions (a0). This is an obvious simplification, especially given the last author's recent models discussing the very different roles of REM vs NREM. Could the authors suggest how different sleep stages may relate to the model or how it could be developed to interact with other successful models such as the ones the last author has developed (e.g., C-HORSE)?
We appreciate the encouragement to comment on the roles of different sleep stages in the manuscript, especially since, as noted, the lab is very interested in this and has explored it in other work. We chose to focus on NREM in this work because the vast majority of electrophysiological studies of sleep replay have identified these events during NREM. In addition, our lab’s theory of the role of REM (Singh et al., 2022, PNAS) is that it is a time for the neocortex to replay remote memories, in complement to the more recent memories replayed during NREM. The experiments we simulate all involve recent memories. Indeed, our view is that part of the reason that there is so little data on REM replay may be that experimenters are almost always looking for traces of recent memories (for good practical and technical reasons).
Regarding the simplicity of the distinction between simulated wake and sleep replay, we view it as an asset of the model that it can account for many of the different characteristics of awake and NREM replay with very simple assumptions about differences in the initial conditions. There are of course many other differences between the states that could be relevant to the impact of replay, but the current target empirical data did not necessitate us taking those into account. This allows us to argue that differences in initial conditions should play a substantial role in an account of the differences between wake and sleep replay.
We have added discussion of these ideas and how they might be incorporated into future versions of the model in the Discussion section:
“Our current simulations have focused on NREM, since the vast majority of electrophysiological studies of sleep replay have identified replay events in this stage. We have proposed in other work that replay during REM sleep may provide a complementary role to NREM sleep, allowing neocortical areas to reinstate remote, already-consolidated memories that need to be integrated with the memories that were recently encoded in the hippocampus and replayed during NREM (Singh et al., 2022). An extension of our model could undertake this kind of continual learning setup, where the student but not teacher network retains remote memories, and the driver of replay alternates between hippocampus (NREM) and cortex (REM) over the course of a night of simulated sleep. Other differences between stages of sleep and between sleep and wake states are likely to become important for a full account of how replay impacts memory. Our current model parsimoniously explains a range of differences between awake and sleep replay by assuming simple differences in initial conditions, but we expect many more characteristics of these states (e.g., neural activity levels, oscillatory profiles, neurotransmitter levels, etc.) will be useful to incorporate in the future.”
Finally, I wonder how the model would explain findings (including the authors') showing a preference for reactivation of weaker memories. The literature seems to suggest that it isn't just a matter of novelty or exposure, but encoding strength. Can the model explain this? Or would it require additional assumptions or some mechanism for selective endogenous reactivation during sleep and rest?
We appreciate the encouragement to discuss this, as we do think the model could explain findings showing a preference for reactivation of weaker memories, as in Schapiro et al. (2018). In our framework, memory strength is reflected in the magnitude of each memory’s associated synaptic weights, so that stronger memories yield higher retrieved‑context activity during wake encoding than weaker ones. Because the model’s suppression mechanism reduces an item’s replay probability in proportion to its retrieved‑context activity, items with larger weights (strong memories) are more heavily suppressed at the onset of replay, while those with smaller weights (weaker memories) receive less suppression. When items have matched reward exposure, this dynamic would bias offline replay toward weaker memories, therefore preferentially reactivating weak memories.
In the section titled “The influence of experience”, we updated a sentence to discuss this idea more explicitly:
“Such a suppression mechanism may be adaptive, allowing replay to benefit not only the most recently or strongly encoded items but also to provide opportunities for the consolidation of weaker or older memories, consistent with empirical evidence (e.g., Schapiro et al. 2018; Yu et al., 2024).”
(5) Lines 186-200 - Perhaps I'm misunderstanding, but wouldn't it be trivial that an external cue at the end-item of Figure 7a would result in backward replay, simply because there is no potential for forward replay for sequences starting at the last item (there simply aren't any subsequent items)? The opposite is true, of course, for the first-item replay, which can't go backward. More generally, my understanding of the literature on forward vs backward replay is that neither is linked to the rodent's location. Both commonly happen at a resting station that is further away from the track. It seems as though the model's result may not hold if replay occurs away from the track (i.e. if a0 would be equal for both pre- and post-run).
In studies where animals run back and forth on a linear track, replay events are decoded separately for left and right runs, identifying both forward and reverse sequences for each direction, for example using direction-specific place cell sequence templates. Accordingly, in our simulation of, e.g., Ambrose et al. (2016), we use two independent sequences, one for left runs and one for right runs (an approach that has been taken in prior replay modeling work). Crucially, our model assumes a context reset between running episodes, preventing the final item of one traversal from acquiring contextual associations with the first item of the next. As a result, learning in the two sequences remains independent, and when an external cue is presented at the track’s end, replay predominantly unfolds in the backward direction, only occasionally producing forward segments when the cue briefly reactivates an earlier sequence item before proceeding forward.
We added a note to the section titled “The context-dependency of memory replay” to clarify this:
“In our model, these patterns are identical to those in our simulation of Ambrose et al. (2016), which uses two independent sequences to mimic the two run directions. This is because the drifting context resets before each run sequence is encoded, with the pause between runs acting as an event boundary that prevents the final item of one traversal from associating with the first item of the next, thereby keeping learning in each direction independent.”
To our knowledge, no study has observed a similar asymmetry when animals are fully removed from the track, although both types of replay can be observed when animals are away from the track. For example, Gupta et al. (2010) demonstrated that when animals replay trajectories far from their current location, the ratio of forward vs. backward replay appears more balanced. We now highlight this result in the manuscript and explain how it aligns with the predictions of our model:
“For example, in tasks where the goal is positioned in the middle of an arm rather than at its end, CMR-replay predicts a more balanced ratio of forward and reverse replay, whereas the EVB model still predicts a dominance of reverse replay due to backward gain propagation from the reward. This contrast aligns with empirical findings showing that when the goal is located in the middle of an arm, replay events are more evenly split between forward and reverse directions (Gupta et al., 2010), whereas placing the goal at the end of a track produces a stronger bias toward reverse replay (Diba & Buzsaki 2007).”
Although no studies, to our knowledge, have observed a context-dependent asymmetry between forward and backward replay when the animal is away from the track, our model does posit conditions under which it could. Specifically, it predicts that deliberation on a specific memory, such as during planning, could generate an internal context input that biases replay: actively recalling the first item of a sequence may favor forward replay, while thinking about the last item may promote backward replay, even when the individual is physically distant from the track.
We now discuss this prediction in the section titled “The context-dependency of memory replay”:
“Our model also predicts that deliberation on a specific memory, such as during planning, could serve to elicit an internal context cue that biases replay: actively recalling the first item of a sequence may favor forward replay, while thinking about the last item may promote backward replay, even when the individual is physically distant from the track. While not explored here, this mechanism presents a potential avenue for future modeling and empirical work.”
(6) The manuscript describes a study by Bendor & Wilson (2012) and tightly mimics their results. However, notably, that study did not find triggered replay immediately following sound presentation, but rather a general bias toward reactivation of the cued sequence over longer stretches of time. In other words, it seems that the model's results don't fully mirror the empirical results. One idea that came to mind is that perhaps it is the R/L context - not the first R/L item - that is cued in this study. This is in line with other TMR studies showing what may be seen as contextual reactivation. If the authors think that such a simulation may better mirror the empirical results, I encourage them to try. If not, however, this limitation should be discussed.
Although our model predicts that replay is triggered immediately by the sound cue, it also predicts a sustained bias toward the cued sequence. Replay in our model unfolds across the rest phase as multiple successive events, so the bias observed in our sleep simulations indeed reflects a prolonged preference for the cued sequence.
We now discuss this issue, acknowledging the discrepancy:
“Bendor and Wilson (2012) found that sound cues during sleep did not trigger immediate replay, but instead biased reactivation toward the cued sequence over an extended period of time. While the model does exhibit some replay triggered immediately by the cue, it also captures the sustained bias toward the cued sequence over an extended period.”
Second, within this framework, context is modeled as a weighted average of the features associated with items. As a result, cueing the model with the first R/L item produces qualitatively similar outcomes as cueing it with a more extended R/L cue that incorporates features of additional items. This is because both approaches ultimately use context features unique to the two sides.
(7) There is some discussion about replay's benefit to memory. One point of interest could be whether this benefit changes between wake and sleep. Relatedly, it would be interesting to see whether the proportion of forward replay, backward replay, or both correlated with memory benefits. I encourage the authors to extend the section on the function of replay and explore these questions.
We thank the reviewer for this suggestion. Regarding differences in the contribution of wake and sleep to memory, our current simulations predict that compared to rest in the task environment, sleep is less biased toward initiating replay at specific items, leading to a more uniform benefit across all memories. Regarding the contributions of forward and backward replay, our model predicts that both strengthen bidirectional associations between items and contexts, benefiting memory in qualitatively similar ways. Furthermore, we suggest that the offline learning captured by our teacher-student simulations reflects consolidation processes that are specific to sleep.
We have expanded the section titled “The influence of experience” to discuss these predictions of the model:
“The results outlined above arise from the model's assumption that replay strengthens bidirectional associations between items and contexts to benefit memory. This assumption leads to several predictions about differences across replay types. First, the model predicts that sleep yields different memory benefits compared to rest in the task environment: Sleep is less biased toward initiating replay at specific items, resulting in a more uniform benefit across all memories. Second, the model predicts that forward and backward replay contribute to memory in qualitatively similar ways but tend to benefit different memories. This divergence arises because forward and backward replay exhibit distinct item preferences, with backward replay being more likely to include rewarded items, thereby preferentially benefiting those memories.”
We also updated the “The function of replay” section to include our teacher-student speculation:
“We speculate that the offline learning observed in these simulations corresponds to consolidation processes that operate specifically during sleep, when hippocampal-neocortical dynamics are especially tightly coupled (Klinzing et al., 2019).”
(8) Replay has been mostly studied in rodents, with few exceptions, whereas CMR and similar models have mostly been used in humans. Although replay is considered a good model of episodic memory, it is still limited due to limited findings of sequential replay in humans and its reliance on very structured and inherently autocorrelated items (i.e., place fields). I'm wondering if the authors could speak to the implications of those limitations on the generalizability of their model. Relatedly, I wonder if the model could or does lead to generalization to some extent in a way that would align with the complementary learning systems framework.
We appreciate these insightful comments. Traditionally, replay studies have focused on spatial tasks with autocorrelated item representations (e.g., place fields). However, an increasing number of human studies have demonstrated sequential replay using stimuli with distinct, unrelated representations. Our model is designed to accommodate both scenarios. In our current simulations, we employ orthogonal item representations while leveraging a shared, temporally autocorrelated context to link successive items. We anticipate that incorporating autocorrelated item representations would further enhance sequence memory by increasing the similarity between successive contexts. Overall, we believe that the model generalizes across a broad range of experimental settings, regardless of the degree of autocorrelation between items. Moreover, the underlying framework has been successfully applied to explain sequential memory in both spatial domains, explaining place cell firing properties (e.g., Howard et al., 2004), and in non-spatial domains, such as free recall experiments where items are arbitrarily related.
In the section titled “A context model of memory replay”, we added this comment to address this point:
“Its contiguity bias stems from its use of shared, temporally autocorrelated context to link successive items, despite the orthogonal nature of individual item representations. This bias would be even stronger if items had overlapping representations, as observed in place fields.”
Since CMR-replay learns distributed context representations where overlap across context vectors captures associative structure, and replay helps strengthen that overlap, this could indeed be viewed as consonant with complementary learning systems integration processes.
Reviewer #2 (Public Review):
This manuscript proposes a model of replay that focuses on the relation between an item and its context, without considering the value of the item. The model simulates awake learning, awake replay, and sleep replay, and demonstrates parallels between memory phenomenon driven by encoding strength, replay of sequence learning, and activation of nearest neighbor to infer causality. There is some discussion of the importance of suppression/inhibition to reduce activation of only dominant memories to be replayed, potentially boosting memories that are weakly encoded. Very nice replications of several key replay findings including the effect of reward and remote replay, demonstrating the equally salient cue of context for offline memory consolidation.
I have no suggestions for the main body of the study, including methods and simulations, as the work is comprehensive, transparent, and well-described. However, I would like to understand how the CMRreplay model fits with the current understanding of the importance of excitation vs inhibition, remembering vs forgetting, activation vs deactivation, strengthening vs elimination of synapses, and even NREM vs REM as Schapiro has modeled. There seems to be a strong association with the efforts of the model to instantiate a memory as well as how that reinstantiation changes across time. But that is not all this is to consolidation. The specific roles of different brain states and how they might change replay is also an important consideration.
We are gratified that the reviewer appreciated the work, and we agree that the paper would benefit from comment on the connections to these other features of consolidation.
Excitation vs. inhibition: CMR-replay does not model variations in the excitation-inhibition balance across brain states (as in other models, e.g., Chenkov et al., 2017), since it does not include inhibitory connections. However, we posit that the experience-dependent suppression mechanism in the model might, in the brain, involve inhibitory processes. Supporting this idea, studies have observed increased inhibition with task repetition (Berners-Lee et al., 2022). We hypothesize that such mechanisms may underlie the observed inverse relationship between task experience and replay frequency in many studies. We discuss this in the section titled “A context model of memory replay”:
“The proposal that a suppression mechanism plays a role in replay aligns with models that regulate place cell reactivation via inhibition (Malerba et al., 2016) and with empirical observations of increased hippocampal inhibitory interneuron activity with experience (Berners-Lee et al., 2022). Our model assumes the presence of such inhibitory mechanisms but does not explicitly model them.”
Remembering/forgetting, activation/deactivation, and strengthening/elimination of synapses: The model does not simulate synaptic weight reduction or pruning, so it does not forget memories through the weakening of associated weights. However, forgetting can occur when a memory is replayed less frequently than others, leading to reduced activation of that memory compared to its competitors during context-driven retrieval. In the Discussion section, we acknowledge that a biologically implausible aspect of our model is that it implements only synaptic strengthening:
“Aspects of the model, such as its lack of regulation of the cumulative positive weight changes that can accrue through repeated replay, are biologically implausible (as biological learning results in both increases and decreases in synaptic weights) and limit the ability to engage with certain forms of low level neural data (e.g., changes in spine density over sleep periods; de Vivo et al., 2017; Maret et al., 2011). It will be useful for future work to explore model variants with more elements of biological plausibility.” Different brain states and NREM vs REM: Reviewer 1 also raised this important issue (see above). We have added the following thoughts on differences between these states and the relationship to our prior work to the Discussion section:
“Our current simulations have focused on NREM, since the vast majority of electrophysiological studies of sleep replay have identified replay events in this stage. We have proposed in other work that replay during REM sleep may provide a complementary role to NREM sleep, allowing neocortical areas to reinstate remote, already-consolidated memories that need to be integrated with the memories that were recently encoded in the hippocampus and replayed during NREM (Singh et al., 2022). An extension of our model could undertake this kind of continual learning setup, where the student but not teacher network retains remote memories, and the driver of replay alternates between hippocampus (NREM) and cortex (REM) over the course of a night of simulated sleep. Other differences between stages of sleep and between sleep and wake states are likely to become important for a full account of how replay impacts memory. Our current model parsimoniously explains a range of differences between awake and sleep replay by assuming simple differences in initial conditions, but we expect many more characteristics of these states (e.g., neural activity levels, oscillatory profiles, neurotransmitter levels, etc.) will be useful to incorporate in the future.”
We hope these points clarify the model’s scope and its potential for future extensions.
Do the authors suggest that these replay systems are more universal to offline processes beyond episodic memory? What about procedural memories and working memory?
We thank the reviewer for raising this important question. We have clarified in the manuscript:
“We focus on the model as a formulation of hippocampal replay, capturing how the hippocampus may replay past experiences through simple and interpretable mechanisms.”
With respect to other forms of memory, we now note that:
“This motor memory simulation using a model of hippocampal replay is consistent with evidence that hippocampal replay can contribute to consolidating memories that are not hippocampally dependent at encoding (Schapiro et al., 2019; Sawangjit et al., 2018). It is possible that replay in other, more domain-specific areas could also contribute (Eichenlaub et al., 2020).”
Though this is not a biophysical model per se, can the authors speak to the neuromodulatory milieus that give rise to the different types of replay?
Our work aligns with the perspective proposed by Hasselmo (1999), which suggests that waking and sleep states differ in the degree to which hippocampal activity is driven by external inputs. Specifically, high acetylcholine levels during waking bias activity to flow into the hippocampus, while low acetylcholine levels during sleep allow hippocampal activity to influence other brain regions. Consistent with this view, our model posits that wake replay is more biased toward items associated with the current resting location due to the presence of external input during waking states. In the Discussion section, we have added a comment on this point:
“Our view aligns with the theory proposed by Hasselmo (1999), which suggests that the degree of hippocampal activity driven by external inputs differs between waking and sleep states: High acetylcholine levels during wakefulness bias activity into the hippocampus, while low acetylcholine levels during slow-wave sleep allow hippocampal activity to influence other brain regions.”
Reviewer #3 (Public Review):
In this manuscript, Zhou et al. present a computational model of memory replay. Their model (CMR-replay) draws from temporal context models of human memory (e.g., TCM, CMR) and claims replay may be another instance of a context-guided memory process. During awake learning, CMR replay (like its predecessors) encodes items alongside a drifting mental context that maintains a recency-weighted history of recently encoded contexts/items. In this way, the presently encoded item becomes associated with other recently learned items via their shared context representation - giving rise to typical effects in recall such as primacy, recency, and contiguity. Unlike its predecessors, CMR-replay has built-in replay periods. These replay periods are designed to approximate sleep or wakeful quiescence, in which an item is spontaneously reactivated, causing a subsequent cascade of item-context reactivations that further update the model's item-context associations.
Using this model of replay, Zhou et al. were able to reproduce a variety of empirical findings in the replay literature: e.g., greater forward replay at the beginning of a track and more backward replay at the end; more replay for rewarded events; the occurrence of remote replay; reduced replay for repeated items, etc. Furthermore, the model diverges considerably (in implementation and predictions) from other prominent models of replay that, instead, emphasize replay as a way of predicting value from a reinforcement learning framing (i.e., EVB, expected value backup).
Overall, I found the manuscript clear and easy to follow, despite not being a computational modeller myself. (Which is pretty commendable, I'd say). The model also was effective at capturing several important empirical results from the replay literature while relying on a concise set of mechanisms - which will have implications for subsequent theory-building in the field.
With respect to weaknesses, additional details for some of the methods and results would help the readers better evaluate the data presented here (e.g., explicitly defining how the various 'proportion of replay' DVs were calculated).
For example, for many of the simulations, the y-axis scale differs from the empirical data despite using comparable units, like the proportion of replay events (e.g., Figures 1B and C). Presumably, this was done to emphasize the similarity between the empirical and model data. But, as a reader, I often found myself doing the mental manipulation myself anyway to better evaluate how the model compared to the empirical data. Please consider using comparable y-axis ranges across empirical and simulated data wherever possible.
We appreciate this point. As in many replay modeling studies, our primary goal is to provide a qualitative fit that demonstrates the general direction of differences between our model and empirical data, without engaging in detailed parameter fitting for a precise quantitative fit. Still, we agree that where possible, it is useful to better match the axes. We have updated figures 2B and 2C so that the y-axis scales are more directly comparable between the empirical and simulated data.
In a similar vein to the above point, while the DVs in the simulations/empirical data made intuitive sense, I wasn't always sure precisely how they were calculated. Consider the "proportion of replay" in Figure 1A. In the Methods (perhaps under Task Simulations), it should specify exactly how this proportion was calculated (e.g., proportions of all replay events, both forwards and backwards, combining across all simulations from Pre- and Post-run rest periods). In many of the examples, the proportions seem to possibly sum to 1 (e.g., Figure 1A), but in other cases, this doesn't seem to be true (e.g., Figure 3A). More clarity here is critical to help readers evaluate these data. Furthermore, sometimes the labels themselves are not the most informative. For example, in Figure 1A, the y-axis is "Proportion of replay" and in 1C it is the "Proportion of events". I presumed those were the same thing - the proportion of replay events - but it would be best if the axis labels were consistent across figures in this manuscript when they reflect the same DV.
We appreciate these useful suggestions. We have revised the Methods section to explain in detail how DVs are calculated for each simulation. The revisions clarify the differences between related measures, such as those shown in Figures 1A and 1C, so that readers can more easily see how the DVs are defined and interpreted in each case.
Reviewer #4/Reviewing Editor (Public Review):
Summary:
With their 'CMR-replay' model, Zhou et al. demonstrate that the use of spontaneous neural cascades in a context-maintenance and retrieval (CMR) model significantly expands the range of captured memory phenomena.
Strengths:
The proposed model compellingly outperforms its CMR predecessor and, thus, makes important strides towards understanding the empirical memory literature, as well as highlighting a cognitive function of replay.
Weaknesses:
Competing accounts of replay are acknowledged but there are no formal comparisons and only CMR-replay predictions are visualized. Indeed, other than the CMR model, only one alternative account is given serious consideration: A variant of the 'Dyna-replay' architecture, originally developed in the machine learning literature (Sutton, 1990; Moore & Atkeson, 1993) and modified by Mattar et al (2018) such that previously experienced event-sequences get replayed based on their relevance to future gain. Mattar et al acknowledged that a realistic Dyna-replay mechanism would require a learned representation of transitions between perceptual and motor events, i.e., a 'cognitive map'. While Zhou et al. note that the CMR-replay model might provide such a complementary mechanism, they emphasize that their account captures replay characteristics that Dyna-replay does not (though it is unclear to what extent the reverse is also true).
We thank the reviewer for these thoughtful comments and appreciate the opportunity to clarify our approach. Our goal in this work is to contrast two dominant perspectives in replay research: replay as a mechanism for learning reward predictions and replay as a process for memory consolidation. These models were chosen as representatives of their classes of models because they use simple and interpretable mechanisms that can simulate a wide range of replay phenomena, making them ideal for contrasting these two perspectives.
Although we implemented CMR-replay as a straightforward example of the memory-focused view, we believe the proposed mechanisms could be extended to other architectures, such as recurrent neural networks, to produce similar results. We now discuss this possibility in the revised manuscript (see below). However, given our primary goal of providing a broad and qualitative contrast of these two broad perspectives, we decided not to undertake simulations with additional individual models for this paper.
Regarding the Mattar & Daw model, it is true that a mechanistic implementation would require a mechanism that avoids precomputing priorities before replay. However, the "need" component of their model already incorporates learned expectations of transitions between actions and events. Thus, the model's limitations are not due to the absence of a cognitive map.
In contrast, while CMR-replay also accumulates memory associations that reflect experienced transitions among events, it generates several qualitatively distinct predictions compared to the Mattar & Daw model. As we note in the manuscript, these distinctions make CMR-replay a contrasting rather than complementary perspective.
Another important consideration, however, is how CMR replay compares to alternative mechanistic accounts of cognitive maps. For example, Recurrent Neural Networks are adept at detecting spatial and temporal dependencies in sequential input; these networks are being increasingly used to capture psychological and neuroscientific data (e.g., Zhang et al, 2020; Spoerer et al, 2020), including hippocampal replay specifically (Haga & Fukai, 2018). Another relevant framework is provided by Associative Learning Theory, in which bidirectional associations between static and transient stimulus elements are commonly used to explain contextual and cue-based phenomena, including associative retrieval of absent events (McLaren et al, 1989; Harris, 2006; Kokkola et al, 2019). Without proper integration with these modeling approaches, it is difficult to gauge the innovation and significance of CMR-replay, particularly since the model is applied post hoc to the relatively narrow domain of rodent maze navigation.
First, we would like to clarify our principal aim in this work is to characterize the nature of replay, rather than to model cognitive maps per se. Accordingly, CMR‑replay is not designed to simulate head‐direction signals, perform path integration, or explain the spatial firing properties of neurons during navigation. Instead, it focuses squarely on sequential replay phenomena, simulating classic rodent maze reactivation studies and human sequence‐learning tasks. These simulations span a broad array of replay experimental paradigms to ensure extensive coverage of the replay findings reported across the literature. As such, the contribution of this work is in explaining the mechanisms and functional roles of replay, and demonstrating that a model that employs simple and interpretable memory mechanisms not only explains replay phenomena traditionally interpreted through a value-based lens but also accounts for findings not addressed by other memory-focused models.
As the reviewer notes, CMR-replay shares features with other memory-focused models. However, to our knowledge, none of these related approaches have yet captured the full suite of empirical replay phenomena, suggesting the combination of mechanisms employed in CMR-replay is essential for explaining these phenomena. In the Discussion section, we now discuss the similarities between CMR-replay and related memory models and the possibility of integrating these approaches:
“Our theory builds on a lineage of memory-focused models, demonstrating the power of this perspective in explaining phenomena that have often been attributed to the optimization of value-based predictions. In this work, we focus on CMR-replay, which exemplifies the memory-centric approach through a set of simple and interpretable mechanisms that we believe are broadly applicable across memory domains. Elements of CMR-replay share similarities with other models that adopt a memory-focused perspective. The model learns distributed context representations whose overlaps encodes associations among items, echoing associative learning theories in which overlapping patterns capture stimulus similarity and learned associations (McLaren & Mackintosh 2002). Context evolves through bidirectional interactions between items and their contextual representations, mirroring the dynamics found in recurrent neural networks (Haga & Futai 2018; Levenstein et al., 2024). However, these related approaches have not been shown to account for the present set of replay findings and lack mechanisms—such as reward-modulated encoding and experience-dependent suppression—that our simulations suggest are essential for capturing these phenomena. While not explored here, we believe these mechanisms could be integrated into architectures like recurrent neural networks (Levenstein et al., 2024) to support a broader range of replay dynamics.”
Recommendations For The Authors
Reviewer #1 (Recommendations For The Authors):
(1) Lines 94-96: These lines may be better positioned earlier in the paragraph.
We now introduce these lines earlier in the paragraph.
(2) Line 103 - It's unclear to me what is meant by the statement that "the current context contains contexts associated with previous items". I understand why a slowly drifting context will coincide and therefore link with multiple items that progress rapidly in time, so multiple items will be linked to the same context and each item will be linked to multiple contexts. Is that the idea conveyed here or am I missing something? I'm similarly confused by line 129, which mentions that a context is updated by incorporating other items' contexts. How could a context contain other contexts?
In the model, each item has an associated context that can be retrieved via Mfc. This is true even before learning, since Mfc is initialized as an identity matrix. During learning and replay, we have a drifting context c that is updated each time an item is presented. At each timestep, the model first retrieves the current item’s associated context cf by Mfc, and incorporates it into c. Equation #2 in the Methods section illustrates this procedure in detail. Because of this procedure, the drifting context c is a weighted sum of past items’ associated contexts.
We recognize that these descriptions can be confusing. We have updated the Results section to better distinguish the drifting context from items’ associated context. For example, we note that:
“We represent the drifting context during learning and replay with c and an item's associated context with cf.”
We have also updated our description of the context drift procedure to distinguish these two quantities:
“During awake encoding of a sequence of items, for each item f, the model retrieves its associated context cf via Mfc. The drifting context c incorporates the item's associated context cf and downweights its representation of previous items' associated contexts (Figure 1c). Thus, the context layer maintains a recency weighted sum of past and present items' associated contexts.”
(3) Figure 1b and 1d - please clarify which axis in the association matrices represents the item and the context.
We have added labels to show what the axes represent in Figure 1.
(4) The terms "experience" and "item" are used interchangeably and it may be best to stick to one term.
We now use the term “item” wherever we describe the model results.
(5) The manuscript describes Figure 6 ahead of earlier figures - the authors may want to reorder their figures to improve readability.
We appreciate this suggestion. We decided to keep the current figure organization since it allows us to group results into different themes and avoid redundancy.
(6) Lines 662-664 are repeated with a different ending, this is likely an error.
We have fixed this error.
Reviewer #3 (Recommendations For The Authors):
Below, I have outlined some additional points that came to mind in reviewing the manuscript - in no particular order.
(1) Figure 1: I found the ordering of panels a bit confusing in this figure, as the reading direction changes a couple of times in going from A to F. Would perhaps putting panel C in the bottom left corner and then D at the top right, with E and F below (also on the right) work?
We agree that this improves the figure. We have restructured the ordering of panels in this figure.
(2) Simulation 1: When reading the intro/results for the first simulation (Figure 2a; Diba & Buszaki, 2007; "When animals traverse a linear track...", page 6, line 186). It wasn't clear to me why pre-run rest would have any forward replay, particularly if pre-run implied that the animal had no experience with the track yet. But in the Methods this becomes clearer, as the model encodes the track eight times prior to the rest periods. Making this explicit in the text would make it easier to follow. Also, was there any reason why specifically eight sessions of awake learning, in particular, were used?
We now make more explicit that the animals have experience with the track before pre-run rest recording:
“Animals first acquire experience with a linear track by traversing it to collect a reward. Then, during the pre-run rest recording, forward replay predominates.”
We included eight sessions of awake learning to match with the number of sessions in Shin et al. (2017), since this simulation attempts to explain data from that study. After each repetition, the model engages in rest. We have revised the Methods section to indicate the motivation for this choice:
“In the simulation that examines context-dependent forward and backward replay through experience (Figs. 2a and 5a), CMR-replay encodes an input sequence shown in Fig. 7a, which simulates a linear track run with no ambiguity in the direction of inputs, over eight awake learning sessions (as in Shin et al. 2019)”
(3) Frequency of remote replay events: In the simulation based on Gupta et al, how frequently overall does remote replay occur? In the main text, the authors mention the mean frequency with which shortcut replay occurs (i.e., the mean proportion of replay events that contain a shortcut sequence = 0.0046), which was helpful. But, it also made me wonder about the likelihood of remote replay events. I would imagine that remote replay events are infrequent as well - given that it is considerably more likely to replay sequences from the local track, given the recency-weighted mental context. Reporting the above mean proportion for remote and local replay events would be helpful context for the reader.
In Figure 4c, we report the proportion of remote replay in the two experimental conditions of Gupta et al. that we simulate.
(4) Point of clarification re: backwards replay: Is backwards replay less likely to occur than forward replay overall because of the forward asymmetry associated with these models? For example, for a backwards replay event to occur, the context would need to drift backwards at least five times in a row, in spite of a higher probability of moving one step forward at each of those steps. Am I getting that right?
The reviewer’s interpretation is correct: CMR-replay is more likely to produce forward than backward replay in sleep because of its forward asymmetry. We note that this forward asymmetry leads to high likelihood of forward replay in the section titled “The context-dependency of memory replay”:
“As with prior retrieved context models (Howard & Kahana 2002; Polyn et al., 2009), CMR-replay encodes stronger forward than backward associations. This asymmetry exists because, during the first encoding of a sequence, an item's associated context contributes only to its ensuing items' encoding contexts. Therefore, after encoding, bringing back an item's associated context is more likely to reactivate its ensuing than preceding items, leading to forward asymmetric replay (Fig. 6d left).”
(5) On terminating a replay period: "At any t, the replay period ends with a probability of 0.1 or if a task-irrelevant item is reactivated." (Figure 1 caption; see also pg 18, line 635). How was the 0.1 decided upon? Also, could you please add some detail as to what a 'task-irrelevant item' would be? From what I understood, the model only learns sequences that represent the points in a track - wouldn't all the points in the track be task-relevant?
This value was arbitrarily chosen as a small value that allows probabilistic stopping. It was not motivated by prior modeling or a systematic search. We have added: “At each timestep, the replay period ends either with a stop probability of 0.1 or if a task-irrelevant item becomes reactivated. (The choice of the value 0.1 was arbitrary; future work could explore the implications of varying this parameter).”
In addition, we now explain in the paper that task irrelevant items “do not appear as inputs during awake encoding, but compete with task-relevant items for reactivation during replay, simulating the idea that other experiences likely compete with current experiences during periods of retrieval and reactivation.”
(6) Minor typos:
Turn all instances of "nonlocal" into "non-local", or vice versa
"For rest at the end of a run, cexternal is the context associated with the final item in the sequence. For rest at the end of a run, cexternal is the context associated with the start item." (pg 20, line 663) - I believe this is a typo and that the second sentence should begin with "For rest at the START of a run".
We have updated the manuscript to correct these typos.
(7) Code availability: I may have missed it, but it doesn't seem like the code is currently available for these simulations. Including the commented code in a public repository (Github, OSF) would be very useful in this case.
We now include a Github link to our simulation code: https://github.com/schapirolab/CMR-replay.
-
-
-
For example, movements advocating for civil liberties, individual rights, and community solidarity push back against what they see as the dehumanizing and homogenizing effects of the managerial state
I bet Civil Liberties as a concept is not in the average liberal's lexicon these days. The state must force WOC's and gays into everything.
I believe that the Gay stuff is all about population control, it's much more moral than the Nazi's so I am not complaining.
Like Historically it does seem to just popped up and taken over institutions.
The "Empire of Love" as Moldbug calls it.
-
-
news.harvard.edu news.harvard.edu
-
If AI is doing your thinking for you, whether it’s through auto-complete or whether it’s in some more sophisticated ways, as in “I’d let AI write the first draft, and then I’ll just edit it,” that is undercutting your critical thinking and your creativity. You may end up using AI to write a job application letter that is the same as everybody else’s because they’re also using AI, and you may lose the job as a result. You always have to remember that the owl sits on your shoulder and not the other way around.
-
If AI is doing your thinking for you, whether it’s through auto-complete or whether it’s in some more sophisticated ways, as in “I’d let AI write the first draft, and then I’ll just edit it,” that is undercutting your critical thinking and your creativity.
Говорится, что делегирование AI ключевых мыслительных задач (даже с последующим редактированием) подрывает развитие собственного критического мышления и креативности.
-
If AI is doing your thinking for you, whether it’s through auto-complete or whether it’s in some more sophisticated ways, as in “I’d let AI write the first draft, and then I’ll just edit it,” that is undercutting your critical thinking and your creativity. You may end up using AI to write a job application letter that is the same as everybody else’s because they’re also using AI, and you may lose the job as a result. You always have to remember that the owl sits on your shoulder and not the other way around.
-
-
sites.google.com sites.google.com
-
nigga
Just as the previous term, this one is charged with a derogatory connotation, so much as it is preferred to refer to it with the euphemistic expression "N-word", which emcompasses both this word and its -er variant. The phenomenon that allowed the erosion of the -er ending is called r-dropping: the -r is replaced by a schwa (ə), an indistinct vowel pronounced "uh". These terms have always been linked with white supremacy, racism and white power. Even though the two terms may seem synonyms, in fact there is a difference: the -er ending word is strictly connected with the all-encompassing hatred and contempt towards black people, whereas the second one is perceived as a term of endearment when uttered by someone belonging to the Black community. Indeed, starting from the 1980s, the word has underwent a process of reclaiming (also called semantic inversion or looping) which corresponds to "taking a word meant as a slur and reappropriating it as a term of endearment" (https://www.washingtonpost.com/sf/national/2014/11/09/the-n-word-an-entrenched-racial-slur-now-more-prevalent-than-ever/?utm_term=.1590a4928864). This strategy allows the originally oppressive term to be re-semantized (that is, to acquire a new meaning) and used to celebrate the community's unique identity and humanity as "an act of redemption by black folk. The word survives on the conditions that black folks have inscribed for it and nobody else can take that. And it becomes violent when other people try to take it and use it." Indeed, white people "have created the word in the first place, but […] they have lost the power to use it with impunity, they have lost the power to reclaim it." […] "If you understand the history of the word and how it's been used, it's not for white people to use […] So if you're not black you can't do that. You actually can't use the word in the way that we use it. It's not possible, because you're not in that space. So any other usage of it is completely wrong." (https://www.bbc.com/news/stories-53749800). https://www.merriam-webster.com/dictionary/N-word https://share.google/2p6rElVA4Vin0v2cC https://www.dailydot.com/irl/how-not-to-use-the-n-word/
Tags
Annotators
URL
-
-
sites.google.com sites.google.com
-
nigga
Just as the previous term, this one is charged with a derogatory connotation, so much as it is preferred to refer to it with the euphemistic expression "N-word", which emcompasses both this word and its -er variant. The phenomenon that allowed the erosion of the -er ending is called r-dropping: the -r is replaced by a schwa (ə), an indistinct vowel pronounced "uh". These terms have always been linked with white supremacy, racism and white power. Even though the two terms may seem synonyms, in fact there is a difference: the -er ending word is strictly connected with the all-encompassing hatred and contempt towards black people, whereas the second one is perceived as a term of endearment when uttered by someone belonging to the Black community. Indeed, starting from the 1980s, the word has underwent a process of reclaiming (also called semantic inversion or looping) which corresponds to "taking a word meant as a slur and reappropriating it as a term of endearment" (https://www.washingtonpost.com/sf/national/2014/11/09/the-n-word-an-entrenched-racial-slur-now-more-prevalent-than-ever/?utm_term=.1590a4928864). This strategy allows the originally oppressive term to be re-semantized (that is, to acquire a new meaning) and used to celebrate the community's unique identity and humanity as "an act of redemption by black folk. The word survives on the conditions that black folks have inscribed for it and nobody else can take that. And it becomes violent when other people try to take it and use it." Indeed, white people "have created the word in the first place, but […] they have lost the power to use it with impunity, they have lost the power to reclaim it." […] "If you understand the history of the word and how it's been used, it's not for white people to use […] So if you're not black you can't do that. You actually can't use the word in the way that we use it. It's not possible, because you're not in that space. So any other usage of it is completely wrong." (https://www.bbc.com/news/stories-53749800). https://www.merriam-webster.com/dictionary/N-word https://share.google/2p6rElVA4Vin0v2cC https://www.dailydot.com/irl/how-not-to-use-the-n-word/
-
-
hamishcampbell.com hamishcampbell.com
-
This is the norm across many #4opens spaces: a near-total lack of interest in building or maintaining shared paths. It’s a textbook case of right-wing Tragedy of the Commons. Developers show up when it suits them, use the space for their narrow needs, then drift off without contributing to the upkeep. They treat community like free infrastructure – something passive they can extract from – rather than a living, tended path we need.
By def, we're not talking about community then. The behaviour mentioned is that of those who do not think they're part of a bigger whole here. Then by def whatever output is there to just use, as there is no social contract involved. Social asymmetry then is a given, and thus a breakdown of commons.
-
-
-
Limitations and Considerations While SQLite is powerful and versatile, it’s important to understand its limitations:ezstandalone.cmd.push(function () { ezstandalone.showAds(119); }); Concurrency: SQLite uses file-based locking, which can limit concurrent write operations. It’s not suitable for high-concurrency scenarios. Network access: SQLite is designed for local storage and doesn’t provide network access out of the box. User management: SQLite doesn’t have built-in user management or access control features.ezstandalone.cmd.push(function () { ezstandalone.showAds(120); }); Scalability: While SQLite can handle databases up to 140 terabytes, it may not be the best choice for very large datasets or high-traffic applications. Alter table limitations: SQLite has limited support for ALTER TABLE operations compared to other database systems.
Llimitations are low concurrency (not an issue, unless I write from multiple applications/scripts to the same thing) local only/mostly, fine too user management, not an issue, just me scalability, not suited to large amounts. The premise is that I won't have lots of data ALTER TABLE limitations, this may mean rebulds/redesigns as things evolve?
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the original reviews.
Public Reviews:
Reviewer #1 (Public review):
Summary
This is a strong paper that presents a clear advance in multi-animal tracking. The authors introduce an updated version of idtracker.ai that reframes identity assignment as a contrastive learning problem rather than a classification task requiring global fragments. This change leads to gains in speed and accuracy. The method eliminates a known bottleneck in the original system, and the benchmarking across species is comprehensive and well executed. I think the results are convincing and the work is significant.
Strengths
The main strengths are the conceptual shift from classification to representation learning, the clear performance gains, and the fact that the new version is more robust. Removing the need for global fragments makes the software more flexible in practice, and the accuracy and speed improvements are well demonstrated. The software appears thoughtfully implemented, with GUI updates and integration with pose estimators.
Weaknesses
I don't have any major criticisms, but I have identified a few points that should be addressed to improve the clarity and accuracy of the claims made in the paper.
(1) The title begins with "New idtracker.ai," which may not age well and sounds more promotional than scientific. The strength of the work is the conceptual shift to contrastive representation learning, and it might be more helpful to emphasize that in the title rather than branding it as "new."
We considered using “Contrastive idtracker.ai”. However, we thought that readers could then think that we believe they could use both the old idtracker.ai or this contrastive version. But we want to say that the new version is the one to use as it is better in both accuracy and tracking times. We think “New idtracker.ai” communicates better that this version is the version we recommend.
(2) Several technical points regarding the comparison between TRex (a system evaluated in the paper) and idtracker.ai should be addressed to ensure the evaluation is fair and readers are fully informed.
(2.1) Lines 158-160: The description of TRex as based on "Protocol 2 of idtracker.ai" overlooks several key additions in TRex, such as posture image normalization, tracklet subsampling, and the use of uniqueness feedback during training. These features are not acknowledged, and it's unclear whether TRex was properly configured - particularly regarding posture estimation, which appears to have been omitted but isn't discussed. Without knowing the actual parameters used to make comparisons, it's difficult to dassess how the method was evaluated.
We added the information about the key additions of TRex in the section “The new idtracker.ai uses representation learning”, lines 153-157. Posture estimation in TRex was not explicitly used but neither disabled during the benchmark; we clarified this in the last paragraph of “Benchmark of accuracy and tracking time”, lines 492-495.
(2.2) Lines 162-163: The paper implies that TRex gains speed by avoiding Protocol 3, but in practice, idtracker.ai also typically avoids using Protocol 3 due to its extremely long runtime. This part of the framing feels more like a rhetorical contrast than an informative one.
We removed this, see new lines 153-157.
(2.3) Lines 277-280: The contrastive loss function is written using the label l, but since it refers to a pair of images, it would be clearer and more precise to write it as l_{I,J}. This would help readers unfamiliar with contrastive learning understand the formulation more easily.
We added this change in lines 613-620.
(2.4) Lines 333-334: The manuscript states that TRex can fail to track certain videos, but this may be inaccurate depending on how the authors classify failures. TRex may return low uniqueness scores if training does not converge well, but this isn't equivalent to tracking failure. Moreover, the metric reported by TRex is uniqueness, not accuracy. Equating the two could mislead readers. If the authors did compare outputs to human-validated data, that should be stated more explicitly.
We observed TRex crashing without outputting any trajectories on some occasions (Appendix 1—figure 1), and this is what we labeled as “failure”. These failures happened in the most difficult videos of our benchmark, that’s why we treated them the same way as idtracker.ai going to P3. We clarified this in new lines 464-469.
The accuracy measured in our benchmark is not estimated but it is human-validated (see section Computation of tracking accuracy in Appendix 1). Both softwares report some quality estimators at the end of a tracking (“estimated accuracy” for idtracker.ai and "uniqueness” for TRex) but these were not used in the benchmark.
(2.5) Lines 339-341: The evaluation approach defines a "successful run" and then sums the runtime across all attempts up to that point. If success is defined as simply producing any output, this may not reflect how experienced users actually interact with the software, where parameters are iteratively refined to improve quality.
Yes, our benchmark was designed to be agnostic to the different experiences of the user. Also, our benchmark was designed for users that do not inspect the trajectories to choose parameters again not to leave room for potential subjectivity.
(2.6) Lines 344-346: The simulation process involves sampling tracking parameters 10,000 times and selecting the first "successful" run. If parameter tuning is randomized rather than informed by expert knowledge, this could skew the results in favor of tools that require fewer or simpler adjustments. TRex relies on more tunable behavior, such as longer fragments improving training time, which this approach may not capture.
We precisely used the TRex parameter track_max_speed to elongate fragments for optimal tracking. Rather than randomized parameter tuning, we defined the “valid range” for this parameter so that all values in it would produce a decent fragment structure. We used this procedure to avoid worsening those methods that use more parameters.
(2.7) Line 354 onward: TRex was evaluated using two varying parameters (threshold and track_max_speed), while idtracker.ai used only one (intensity_threshold). With a fixed number of samples, this asymmetry could bias results against TRex. In addition, users typically set these parameters based on domain knowledge rather than random exploration.
idtracker.ai and TRex have several parameters. Some of them have a single correct value (e.g. number of animals) or the default value that the system computes is already good (e.g. minimum blob size). For a second type of parameters, the system finds a value that is in general not as good, so users need to modify them. In general, users find that for this second type of parameter there is a valid interval of possible values, from which they need to choose a single value to run the system. idtracker.ai has intensity_threshold as the only parameter of this second type and TRex has two: threshold and track_max_speed. For these parameters, choosing one value or another within the valid interval can give different tracking results. Therefore, when we model a user that wants to run the system once except if it goes to P3 (idtracker.ai) or except if it crashes (TRex), it is these parameters we sample from within the valid interval to get a different value for each run of the system. We clarify this in lines 452-469 of the section “Benchmark of accuracy and tracking time”.
Note that if we chose to simply run old idtracker.ai (v4 or v5) or TRex a single time, this would benefit the new idtracker.ai (v6). This is because old idtracker.ai can enter the very slow protocol 3 and TRex can fail to track. So running old idtracker.ai or TRex up to 5 times until old idtracker.ai does not use Protocol 3 and TRex does not fail is to make them as good as they can be with respect to the new idtracker.ai.
(2.8) Figure 2-figure supplement 3: The memory usage comparison lacks detail. It's unclear whether RAM or VRAM was measured, whether shared or compressed memory was included, or how memory was sampled. Since both tools dynamically adjust to system resources, the relevance of this comparison is questionable without more technical detail.
We modified the text in the caption (new Figure 1-figure supplement 2) adding the kind of memory we measured (RAM) and how we measured it. We already have a disclaimer for this plot saying that memory management depends on the machine's available resources. We agree that this is a simple analysis of the usage of computer resources.
(3) While the authors cite several key papers on contrastive learning, they do not use the introduction or discussion to effectively situate their approach within related fields where similar strategies have been widely adopted. For example, contrastive embedding methods form the backbone of modern facial recognition and other image similarity systems, where the goal is to map images into a latent space that separates identities or classes through clustering. This connection would help emphasize the conceptual strength of the approach and align the work with well-established applications. Similarly, there is a growing literature on animal re-identification (ReID), which often involves learning identity-preserving representations across time or appearance changes. Referencing these bodies of work would help readers connect the proposed method with adjacent areas using similar ideas, and show that the authors are aware of and building on this wider context.
We have now added a new section in Appendix 3, “Differences with previous work in contrastive/metric learning” (lines 792-841) to include references to previous work and a description of what we do differently.
(4) Some sections of the Results text (e.g., lines 48-74) read more like extended figure captions than part of the main narrative. They include detailed explanations of figure elements, sorting procedures, and video naming conventions that may be better placed in the actual figure captions or moved to supplementary notes. Streamlining this section in the main text would improve readability and help the central ideas stand out more clear
Thank you for pointing this out. We have rewritten the Results, for example streamlining the old lines 48-74 (new lines 42-48) by moving the comments about names, files and order of videos to the caption of Figure 1.
Overall, though, this is a high-quality paper. The improvements to idtracker.ai are well justified and practically significant. Addressing the above comments will strengthen the work, particularly by clarifying the evaluation and comparisons.
We thank the reviewer for the detailed suggestions. We believe we have taken all of them into consideration to improve the ms.
Reviewer #2 (Public review):
Summary:
This work introduces a new version of the state-of-the-art idtracker.ai software for tracking multiple unmarked animals. The authors aimed to solve a critical limitation of their previous software, which relied on the existence of "global fragments" (video segments where all animals are simultaneously visible) to train an identification classifier network, in addition to addressing concerns with runtime speed. To do this, the authors have both re-implemented the backend of their software in PyTorch (in addition to numerous other performance optimizations) as well as moving from a supervised classification framework to a self-supervised, contrastive representation learning approach that no longer requires global fragments to function. By defining positive training pairs as different images from the same fragment and negative pairs as images from any two co-existing fragments, the system cleverly takes advantage of partial (but high-confidence) tracklets to learn a powerful representation of animal identity without direct human supervision. Their formulation of contrastive learning is carefully thought out and comprises a series of empirically validated design choices that are both creative and technically sound. This methodological advance is significant and directly leads to the software's major strengths, including exceptional performance improvements in speed and accuracy and a newfound robustness to occlusion (even in severe cases where no global fragments can be detected). Benchmark comparisons show the new software is, on average, 44 times faster (up to 440 times faster on difficult videos) while also achieving higher accuracy across a range of species and group sizes. This new version of idtracker.ai is shown to consistently outperform the closely related TRex software (Walter & Couzin, 2021\), which, together with the engineering innovations and usability enhancements (e.g., outputs convenient for downstream pose estimation), positions this tool as an advancement on the state-of-the-art for multi-animal tracking, especially for collective behavior studies.
Despite these advances, we note a number of weaknesses and limitations that are not well addressed in the present version of this paper:
Weaknesses
(1) The contrastive representation learning formulation. Contrastive representation learning using deep neural networks has long been used for problems in the multi-object tracking domain, popularized through ReID approaches like DML (Yi et al., 2014\) and DeepReID (Li et al., 2014). More recently, contrastive learning has become more popular as an approach for scalable self-supervised representation learning for open-ended vision tasks, as exemplified by approaches like SimCLR (Chen et al., 2020), SimSiam (Chen et al., 2020\), and MAE (He et al., 2021\) and instantiated in foundation models for image embedding like DINOv2 (Oquab et al., 2023). Given their prevalence, it is useful to contrast the formulation of contrastive learning described here relative to these widely adopted approaches (and why this reviewer feels it is appropriate):
(1.1) No rotations or other image augmentations are performed to generate positive examples. These are not necessary with this approach since the pairs are sampled from heuristically tracked fragments (which produces sufficient training data, though see weaknesses discussed below) and the crops are pre-aligned egocentrically (mitigating the need for rotational invariance).
(1.2) There is no projection head in the architecture, like in SimCLR. Since classification/clustering is the only task that the system is intended to solve, the more general "nuisance" image features that this architectural detail normally affords are not necessary here.
(1.3) There is no stop gradient operator like in BYOL (Grill et al., 2020\) or SimSiam. Since the heuristic tracking implicitly produces plenty of negative pairs from the fragments, there is no need to prevent representational collapse due to class asymmetry. Some care is still needed, but the authors address this well through a pair sampling strategy (discussed below).
(1.4) Euclidean distance is used as the distance metric in the loss rather than cosine similarity as in most contrastive learning works. While cosine similarity coupled with L2-normalized unit hypersphere embeddings has proven to be a successful recipe to deal with the curse of dimensionality (with the added benefit of bounded distance limits), the authors address this through a cleverly constructed loss function that essentially allows direct control over the intra- and inter-cluster distance (D\_pos and D\_neg). This is a clever formulation that aligns well with the use of K-means for the downstream assignment step.
No concerns here, just clarifications for readers who dig into the review. Referencing the above literature would enhance the presentation of the paper to align with the broader computer vision literature.
Thank you for this detailed comparison. We have now added a new section in Appendix 3, “Differences with previous work in contrastive/metric learning” (lines 792-841) to include references to previous work and a description of what we do differently, including the points raised by the reviewer.
(2) Network architecture for image feature extraction backbone. As most of the computations that drive up processing time happen in the network backbone, the authors explored a variety of architectures to assess speed, accuracy, and memory requirements. They land on ResNet18 due to its empirically determined performance. While the experiments that support this choice are solid, the rationale behind the architecture selection is somewhat weak. The authors state that: "We tested 23 networks from 8 different families of state-of-the-art convolutional neural network architectures, selected for their compatibility with consumer-grade GPUs and ability to handle small input images (20 × 20 to 100 × 100 pixels) typical in collective animal behavior videos."
(2.1) Most modern architectures have variants that are compatible with consumer-grade GPUs. This is true of, for example, HRNet (Wang et al., 2019), ViT (Dosovitskiy et al., 2020), SwinT (Liu et al., 2021), or ConvNeXt (Liu et al., 2022), all of which report single GPU training and fast runtime speeds through lightweight configuration or subsequent variants, e.g., MobileViT (Mehta et al., 2021). The authors may consider revising that statement or providing additional support for that claim (e.g., empirical experiments) given that these have been reported to outperform ResNet18 across tasks.
Following the recommendation of the reviewer, we tested the architectures SwinT, ConvNeXt and ViT. We found out that none of them outperformed ResNet18 since they all showed a slower learning curve. This would result in higher tracking times. These tests are now included in the section “Network architecture” (lines 550-611).
(2.2) The compatibility of different architectures with small image sizes is configurable. Most convolutional architectures can be readily adapted to work with smaller image sizes, including 20x20 crops. With their default configuration, they lose feature map resolution through repeated pooling and downsampling steps, but this can be readily mitigated by swapping out standard convolutions with dilated convolutions and/or by setting the stride of pooling layers to 1, preserving feature map resolution across blocks. While these are fairly straightforward modifications (and are even compatible with using pretrained weights), an even more trivial approach is to pad and/or resize the crops to the default image size, which is likely to improve accuracy at a possibly minimal memory and runtime cost. These techniques may even improve the performance with the architectures that the authors did test out.
The only two tested architectures that require a minimum image size are AlexNet and DenseNet. DenseNet proved to underperform ResNet18 in the videos where the images are sufficiently large. We have tested AlexNet with padded images to see that it also performs worse than ResNet18 (see Appendix 3—figure 1).
We also tested the initialization of ResNet18 with pre-trained weights from ImageNet (in Appendix 3—figure 2) and it proved to bring no benefit to the training speed (added in lines 591-592).
(2.3) The authors do not report whether the architecture experiments were done with pretrained or randomly initialized weights.
We adapted the text to make it clear that the networks are always randomly initialized (lines 591-592, lines 608-609 and the captions of Appendix 3—figure 1 and 2).
(2.4) The authors do not report some details about their ResNet18 design, specifically whether a global pooling layer is used and whether the output fully connected layer has any activation function. Additionally, they do not report the version of ResNet18 employed here, namely, whether the BatchNorm and ReLU are applied after (v1) or before (v2) the conv layers in the residual path.
We use ResNet18 v1 with no activation function nor bias in its last layer (this has been clarified in the lines 606-608). Also, by design, ResNet has a global average pool right before the last fully connected layer which we did not remove. In response to the reviewer, Resnet18 v2 was tested and its performance is the same as that of v1 (see Appendix 3—figure 1 and lines 590-591).
(3) Pair sampling strategy. The authors devised a clever approach for sampling positive and negative pairs that is tailored to the nature of the formulation. First, since the positive and negative labels are derived from the co-existence of pretracked fragments, selection has to be done at the level of fragments rather than individual images. This would not be the case if one of the newer approaches for contrastive learning were employed, but it serves as a strength here (assuming that fragment generation/first pass heuristic tracking is achievable and reliable in the dataset). Second, a clever weighted sampling scheme assigns sampling weights to the fragments that are designed to balance "exploration and exploitation". They weigh samples both by fragment length and by the loss associated with that fragment to bias towards different and more difficult examples.
(3.1) The formulation described here resembles and uses elements of online hard example mining (Shrivastava et al., 2016), hard negative sampling (Robinson et al., 2020\), and curriculum learning more broadly. The authors may consider referencing this literature (particularly Robinson et al., 2020\) for inspiration and to inform the interpretation of the current empirical results on positive/negative balancing.
Following this recommendation, we added references of hard negative mining in the new section “Differences with previous work in contrastive/metric learning”, lines 792-841. Regarding curriculum learning, even though in spirit it might have parallels with our sampling method in the sense that there is a guided training of the network, we believe the approach is more similar to an exploration-exploitation paradigm.
(4) Speed and accuracy improvements. The authors report considerable improvements in speed and accuracy of the new idTracker (v6) over the original idTracker (v4?) and TRex. It's a bit unclear, however, which of these are attributable to the engineering optimizations (v5?) versus the representation learning formulation.
(4.1) Why is there an improvement in accuracy in idTracker v5 (L77-81)? This is described as a port to PyTorch and improvements largely related to the memory and data loading efficiency. This is particularly notable given that the progression went from 97.52% (v4; original) to 99.58% (v5; engineering enhancements) to 99.92% (v6; representation learning), i.e., most of the new improvement in accuracy owes to the "optimizations" which are not the central emphasis of the systematic evaluations reported in this paper.
V5 was a two year-effort designed to improve time efficiency of v4. It was also a surprise to us that accuracy was higher, but that likely comes from the fact that the substituted code from v4 contained some small bug/s. The improvements in v5 are retained in v6 (contrastive learning) and v6 has higher accuracy and shorter tracking times. The difference in v6 for this extra accuracy and shorter tracking times is contrastive learning.
(4.2) What about the speed improvements? Relative to the original (v4), the authors report average speed-ups of 13.6x in v5 and 44x in v6. Presumably, the drastic speed-up in v6 comes from a lower Protocol 2 failure rate, but v6 is not evaluated in Figure 2 - figure supplement 2.
Idtracker.ai v5 runs an optimized Protocol 2 and, sometimes, the Protocol 3. But v6 doesn’t run either of them. While P2 is still present in v6 as a fallback protocol when contrastive fails, in our v6 benchmark P2 was never needed. So the v6 speedup comes from replacing both P2 and P3 with the contrastive algorithm.
(5) Robustness to occlusion. A major innovation enabled by the contrastive representation learning approach is the ability to tolerate the absence of a global fragment (contiguous frames where all animals are visible) by requiring only co-existing pairs of fragments owing to the paired sampling formulation. While this removes a major limitation of the previous versions of idtracker.ai, its evaluation could be strengthened. The authors describe an ablation experiment where an arc of the arena is masked out to assess the accuracy under artificially difficult conditions. They find that the v6 works robustly up to significant proportions of occlusions, even when doing so eliminates global fragments.
(5.1) The experiment setup needs to be more carefully described.
(5.1.1) What does the masking procedure entail? Are the pixels masked out in the original video or are detections removed after segmentation and first pass tracking is done?
The mask is defined as a region of interest in the software. This means that it is applied at the segmentation step where the video frame is converted to a foreground-background binary image. The region of interest is applied here, converting to background all pixels not inside of it. We clarified this in the newly added section Occlusion tests, lines 240-244.
(5.1.2) What happens at the boundary of the mask? (Partial segmentation masks would throw off the centroids, and doing it after original segmentation does not realistically model the conditions of entering an occlusion area.)
Animals at the boundaries of the mask are partially detected. This can change the location of their detected centroid. That’s why, when computing the ground-truth accuracy for these videos, only the groundtruth centroids that were at minimum 15 pixels further from the mask were considered. We clarified this in the newly added section Occlusion tests, lines 248-251.
(5.1.3) Are fragments still linked for animals that enter and then exit the mask area?
No artificial fragment linking was added in these videos. Detected fragments are linked the usual way. If one animal hides into the mask, the animal disappears so the fragment breaks. We clarified this in the newly added section Occlusion tests, lines 245-247.
(5.1.4) How is the evaluation done? Is it computed with or without the masked region detections?
The groundtruth used to validate these videos contains the positions of all animals at all times. But only the positions outside the mask at each frame were considered to compute the tracking accuracy. We clarified this in the newly added section Occlusion tests, lines 248-251.
(5.2) The circular masking is perhaps not the most appropriate for the mouse data, which is collected in a rectangular arena.
We wanted to show the same proof of concept in different videos. For that reason, we used to cover the arena parametrized by an angle. In the rectangular arena the circular masking uses an external circle, so it is covering the rectangle parametrized by an angle.
(5.3) The number of co-existing fragments, which seems to be the main determinant of performance that the authors derive from this experiment, should be reported for these experiments. In particular, a "number of co-existing fragments" vs accuracy plot would support the use of the 0.25(N-1) heuristic and would be especially informative for users seeking to optimize experimental and cage design. Additionally, the number of co-existing fragments can be artificially reduced in other ways other than a fixed occlusion, including random dropout, which would disambiguate it from potential allocentric positional confounds (particularly relevant in arenas where egocentric pose is correlated with allocentric position).
We included the requested analysis about the fragment connectivity in Figure 3-figure supplement 1. We agree that there can be additional ways of reducing co-existing fragments, but we think the occlusion tests have the additional value that there are many real experiments similar to this test.
(6) Robustness to imaging conditions. The authors state that "the new idtracker.ai can work well with lower resolutions, blur and video compression, and with inhomogeneous light (Figure 2 - figure supplement 4)." (L156). Despite this claim, there are no speed or accuracy results reported for the artificially corrupted data, only examples of these image manipulations in the supplementary figure.
We added this information in the same image, new Figure 1 - figure supplement 3.
(7) Robustness across longitudinal or multi-session experiments. The authors reference idmatcher.ai as a compatible tool for this use case (matching identities across sessions or long-term monitoring across chunked videos), however, no performance data is presented to support its usage. This is relevant as the innovations described here may interact with this setting. While deep metric learning and contrastive learning for ReID were originally motivated by these types of problems (especially individuals leaving and entering the FOV), it is not clear that the current formulation is ideally suited for this use case. Namely, the design decisions described in point 1 of this review are at times at odds with the idea of learning generalizable representations owing to the feature extractor backbone (less scalable), low-dimensional embedding size (less representational capacity), and Euclidean distance metric without hypersphere embedding (possible sensitivity to drift). It's possible that data to support point 6 can mitigate these concerns through empirical results on variations in illumination, but a stronger experiment would be to artificially split up a longer video into shorter segments and evaluate how generalizable and stable the representations learned in one segment are across contiguous ("longitudinal") or discontiguous ("multi-session") segments.
We have now added a test to prove the reliability of idmatcher.ai in v6. In this test, 14 videos are taken from the benchmark and split in two non-overlapping parts (with a 200 frames gap in between). idmatcher.ai is run between the two parts presenting a 100% accuracy identity matching across all of them (see section “Validity of idmatcher.ai in the new idtracker.ai”, lines 969-1008).
We thank the reviewer for the detailed suggestions. We believe we have taken all of them into consideration to improve the ms.
Reviewer #3 (Public review):
Summary
The authors propose a new version of idTracker.ai for animal tracking. Specifically, they apply contrastive learning to embed cropped images of animals into a feature space where clusters correspond to individual animal identities.
Strengths
By doing this, the new software alleviates the requirement for so-called global fragments - segments of the video, in which all entities are visible/detected at the same time - which was necessary in the previous version of the method. In general, the new method reduces the tracking time compared to the previous versions, while also increasing the average accuracy of assigning the identity labels.
Weaknesses
The general impression of the paper is that, in its current form, it is difficult to disentangle the old from the new method and understand the method in detail. The manuscript would benefit from a major reorganization and rewriting of its parts. There are also certain concerns about the accuracy metric and reducing the computational time.
We have made the following modifications in the presentation:
(1) We have added section tiles to the main text so it is clearer what tracking system we are referring to. For example, we now have sections “Limitation of the original idtracker.ai”, “Optimizing idtracker.ai without changes in the learning method” and “The new idtracker.ai uses representation learning”.
(2) We have completely rewritten all the text of the ms until we start with contrastive learning. Old L20-89 is now L20-L66, much shorter and easier to read.
(3) We have rewritten the first 3 paragraphs in the section “The new idtracker.ai uses representation learning” (lines 68-92).
(4) We now expanded Appendix 3 to discuss the details of our approach (lines 539-897). It discusses in detail the steps of the algorithm, the network architecture, the loss function, the sampling strategy, the clustering and identity assignment, and the stopping criteria in training
(5) To cite previous work in detail and explain what we do differently, we have now added in Appendix 3 the new section “Differences with previous work in contrastive/metric learning” (lines 792-841).
Regarding accuracy metrics, we have replaced our accuracy metric with the standard metric IDF1. IDF1 is the standard metric that is applied to systems in which the goal is to maintain consistent identities across time. See also the section in Appendix 1 "Computation of tracking accuracy” (lines 414-436) explaining IDF1 and why this is an appropriate metric for our goal.
Using IDF1 we obtain slightly higher accuracies for the idtracker.ai systems. This is the comparison of mean accuracy over all our benchmark for our previous accuracy score and the new one for the full trajectories:
v4: 97.42% -> 98.24%
v5: 99.41% -> 99.49%
v6: 99.74% -> 99.82%
trex: 97.89% -> 97.89%
We thank the reviewer for the suggestions about presentation and about the use of more standard metrics.
Recommendations for the authors:
Reviewer #2 (Recommendations for the authors):
(1) Figure 1a: A graphical legend inset would make it more readable since there are multiple colors, line styles, and connecting lines to parse out.
Following this recommendation, we added a graphical legend in the old Figure 1 (new Figure 2).
(2) L46: "have images" → "has images".
We applied this correction. Line 35.
(3) L52: "videos start with a letter for the species (z,**f**,m)", but "d" is used for fly videos.
We applied this correction in the caption of Figure 1.
(4) L62: "with Protocol 3 a two-step process" → "with Protocol 3 being a two-step process".
We rewrote this paragraph without mentioning Protocol 3, lines 37-41.
(5) L82-89: This is the main statement of the problems that are being addressed here (speed and relaxing the need for global fragments). This could be moved up, emphasized, and made clearer without the long preamble and results on the engineering optimizations in v5. This lack of linearity in the narrative is also evident in the fact that after Figure 1a is cited, inline citations skip to Figure 2 before returning to Figure 1 once the contrastive learning is introduced.
We have rewritten all the text until the contrastive learning, (old lines 20-89 are now lines 20-66). The text is shorter, more linear and easier to read.
(6) L114: "pairs until the distance D_{pos}" → "pairs until the distance approximates D_{pos}".
We rewrote as “ pairs until the distance 𝐷pos (or 𝐷neg) is reached” in line 107.
(7) L570: Missing a right parenthesis in the equation.
We no longer have this equation in the ms.
(8) L705: "In order to identify fragments we, not only need" → "In order to identify fragments, we not only need".
We applied this correction, Line 775.
(9) L819: "probably distribution" → "probability distribution".
We applied this correction, Line 776.
(10) L833: "produced the best decrease the time required" → "produced the best decrease of the time required".
We applied this correction, Line 746.
Reviewer #3 (Recommendations for the authors):
(1) We recommend rewriting and restructuring the manuscript. The paper includes a detailed explanation of the previous approaches (idTracker and idTracker.ai) and their limitations. In contrast, the description of the proposed method is short and unstructured, which makes it difficult to distinguish between the old and new methods as well as to understand the proposed method in general. Here are a few examples illustrating the problem.
(1.1) Only in line 90 do the authors start to describe the work done in this manuscript. The previous 3 pages list limitations of the original method.
We have now divided the main text into sections, so it is clearer what is the previous method (“Limitation of the original idtracker.ai”, lines 28-51), the new optimization we did of this method (“Optimizing idtracker.ai without changes in the learning method”, lines 52-66) and the new contrastive approach that also includes the optimizations (“The new idtracker.ai uses representation learning”, lines 66-164). Also, the new text has now been streamlined until the contrastive section, following your suggestion. You can see that in the new writing the three sections are 25 , 15 and 99 lines. The more detailed section is the new system, the other two are needed as reference, to describe which problem we are solving and the extra new optimizations.
(1.2) The new method does not have a distinct name, and it is hard to follow which idtracker.ai is a specific part of the text referring to. Not naming the new method makes it difficult to understand.
We use the name new idtracker.ai (v6) so it becomes the current default version. v5 is now obsolete, as well as v4. And from the point of view of the end user, no new name is needed since v6 is just an evolution of the same software they have been using. Also, we added sections in the main text to clarify the ideas in there and indicate the version of idtracker.ai we are referring to.
(1.3) There are "Protocol 2" and "Protocol 3" mixed with various versions of the software scattered throughout the text, which makes it hard to follow. There should be some systematic naming of approaches and a listing of results introduced.
Following this recommendation we no longer talk about the specific protocols of the old version of idtracker.ai in the main text. We rewritten the explanation of these versions in a more clear and straightforward way, lines 29-36.
(2) To this end, the authors leave some important concepts either underexplained or only referenced indirectly via prior work. For example, the explanation of how the fragments are created (line 15) is only explained by the "video structure" and the algorithm that is responsible for resolving the identities during crossings is not detailed (see lines 46-47, 149-150). Including summaries of these elements would improve the paper's clarity and accessibility.
We listed the specific sections from our previous publication where the reader can find information about the entire tracking pipeline (lines 539-549). This way, we keep the ms clear and focused on the new identification algorithm while indicating where to find such information.
(3) Accuracy metrics are not clear. In line 319, the authors define it as based on "proportion of errors in the trajectory". This proportion is not explained. How is the error calculated if a trajectory is lost or there are identity swaps? Multi-object tracking has a range of accuracy metrics that account for such events but none of those are used by the authors. Estimating metrics that are common for MOT literature, for example, IDF1, MOTA, and MOTP, would allow for better method performance understanding and comparison.
In the new ms, we replaced our accuracy metric with the standard metric IDF1. IDF1 is the standard metric that is applied to systems in which the goal is to maintain consistent identities across time. See also the section in Appendix 1 "Computation of tracking accuracy” explaining why IDF1 and not MOTA or MOTP is the adequate metric for a system that wants to give correct tracking by identification in time. See lines 416-436.
Using IDF1 we obtain slightly higher accuracies for the idtracker.ai systems. This is the comparison of mean accuracy four our previous accuracy and the new one for the full trajectories:
v4: 97.42% -> 98.24%
v5: 99.41% -> 99.49%
v6: 99.74% -> 99.82%
trex: 97.89% -> 97.89%
(4) Additionally, the authors distinguish between tracking with and without crossings, but do not provide statistics on the frequency of crossings per video. It is also unclear how the crossings are considered for the final output. Including information such as the frame rate of the videos would help to better understand the temporal resolution and the differences between consecutive frames of the videos.
We added this information in the Appendix 1 “Benchmark of accuracy and tracking time”, lines 445-451. The framerate in our benchmark videos goes from 25 to 60 fps (average of 37 fps). On average 2.6% of the blobs are crossings (1.1% for zebrafish 0.7% for drosophila 9.4% for mice).
(5) In the description of the dataset used for evaluation (lines 349-365), the authors describe the random sampling of parameter values for each tracking run. However, it is unclear whether the same values were used across methods. Without this clarification, comparisons between the proposed method, older versions, and TRex might be biased due to lucky parameter combinations. In addition, the ranges from which the values were randomly sampled were also not described.
Only one parameter is shared between idtracker.ai and TRex: intensity_threshold (in idtracker.ai) and threshold (in TRex). Both are conceptually equivalent but differ in their numerical values since they affect different algorithms. V4, v5, and TRex each required the same process of independent expert visual inspection of the segmentation to select the valid value range. Since versions 5 and 6 use exactly the same segmentation algorithm, they share the same parameter ranges.
All the ranges of valid values used in our benchmark are public here https://drive.google.com/drive/folders/1tFxdtFUudl02ICS99vYKrZLeF28TiYpZ as stated in the section “Data availability”, lines 227-228.
(6) Lines 122-123, Figure 1c. "batches" - is an imprecise metric of training time as there is no information about the batch size.
We clarified the Figure caption, new Figure 2c.
(7) Line 145 - "we run some steps... For example..." leaves the method description somewhat unclear. It would help if you could provide more details about how the assignments are carried out and which metrics are being used.
Following this recommendation, we listed the specific sections from our previous publication where the reader can find information about the entire tracking pipeline (lines 539-549). This way, we keep the ms clear and focused on the new identification algorithm while indicating where to find such information.
(8) Figure 3. How is tracking accuracy assessed with occlusions? Are the individuals correctly recognized when they reappear from the occluded area?
The groundtruth for this video contains the positions of all animals at all times. Only the groundtruth points inside the region of interest are taken into account when computing the accuracy. When the tracking reaches high accuracy, it means that animals are successfully relabeled every time they enter the non-masked region. Note that this software works all the time by identification of animals, so crossings and occlusion are treated the same way. What is new here is that the occlusions are so large that there are no global fragments. We clarified this in the new section “Occlusion tests” in Methods, lines 239-251.
(9) Lines 185-187 this part of the sentence is not clear.
We rewrote this part in a clearer way, lines 180-182.
(10) The authors also highlight the improved runtime performance. However, they do not provide a detailed breakdown of the time spent on each component of the tracking/training pipeline. A timing breakdown would help to compare the training duration with the other components. For example, the calculation of the Silhouette Score alone can be time-consuming and could be a bottleneck in the training process. Including this information would provide a clearer picture of the overall efficiency of the method.
We measured that the training of ResNet takes on average in our benchmark 47% of the tracking time (we added this information line 551 section “Network Architecture”). In this training stage the bottleneck becomes the network forward and backward pass, limited by the GPU performance. All other processes happening during training have been deeply optimized and parallelized when needed so their contribution to the training time is minimal. Apart from the training, we also measured 24.4% of the total tracking time spent in reading and segmenting the video files and 11.1% in processing the identification images and detecting crossings.
(11) An important part of the computational cost is related to model training. It would be interesting to test whether a model trained on one video of a specific animal type (e.g., zebrafish_5) generalizes to another video of the same type (e.g., zebrafish_7). This would assess the model's generalizability across different videos of the same species and spare a lot of compute. Alternatively, instead of training a model from scratch for each video, the authors could also consider training a base model on a superset of images from different videos and then fine-tuning it with a lower learning rate for each specific video. This could potentially save time and resources while still achieving good performance.
Already before v6, there was the possibility for the user to start training the identification network by copying the final weights from another tracking session. This knowledge transfer feature is still present in v6 and it still decreases the training times significatively. This information has been added in Appendix 4, lines 906-909.
We have already begun working on the interesting idea of a general base model but it brings some complex challenges. It could be a very useful new feature for future idtracker.ai releases.
We thank the reviewer for the many suggestions. We have implemented all of them.
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the original reviews.
Public Reviews:
Reviewer #1 (Public review):
Summary:
Artiushin et al. establish a comprehensive 3D atlas of the brain of the orb-web building spider Uloborus diversus. First, they use immunohistochemistry detection of synapsin to mark and reconstruct the neuropils of the brain of six specimens and they generate a standard brain by averaging these brains. Onto this standard 3D brain, they plot immunohistochemical stainings of major transmitters to detect cholinergic, serotonergic, octopaminergic/taryminergic and GABAergic neurons, respectively. Further, they add information on the expression of a number of neuropeptides (Proctolin, AllatostatinA, CCAP, and FMRFamide). Based on this data and 3D reconstructions, they extensively describe the morphology of the entire synganglion, the discernible neuropils, and their neurotransmitter/neuromodulator content.
Strengths:
While 3D reconstruction of spider brains and the detection of some neuroactive substances have been published before, this seems to be the most comprehensive analysis so far, both in terms of the number of substances tested and the ambition to analyze the entire synganglion. Interestingly, besides the previously described neuropils, they detect a novel brain structure, which they call the tonsillar neuropil.<br /> Immunohistochemistry, imaging, and 3D reconstruction are convincingly done, and the data are extensively visualized in figures, schemes, and very useful films, which allow the reader to work with the data. Due to its comprehensiveness, this dataset will be a valuable reference for researchers working on spider brains or on the evolution of arthropod brains.
Weaknesses:
As expected for such a descriptive groundwork, new insights or hypotheses are limited, apart from the first description of the tonsillar neuropil. A more comprehensive labeling in the panels of the mentioned structures would help to follow the descriptions. The reconstruction of the main tracts of the brain would be a very valuable complementary piece of data.
Reviewer #2 (Public review):
Summary
Artiushin et al. created the first three-dimensional atlas of a synganglion in the hackled orb-weaver spider, which is becoming a popular model for web-building behavior. Immunohistochemical analysis with an impressive array of antisera reveals subcompartments of neuroanatomical structures described in other spider species as well as two previously undescribed arachnid structures, the protocerebral bridge, hagstone, and paired tonsillar neuropils. The authors describe the spider's neuroanatomy in detail and discuss similarities and differences from other spider species. The final section of the discussion examines the homology between onychophoran and chelicerate arcuate bodies and mandibulate central bodies.
Strengths
The authors set out to create a detailed 3D atlas and accomplished this goal.
Exceptional tissue clearing and imaging of the nervous system reveal the three-dimensional relationships between neuropils and some connectivity that would not be apparent in sectioned brains.
A detailed anatomical description makes it easy to reference structures described between the text and figures.
The authors used a large palette of antisera which may be investigated in future studies for function in the spider nervous system and may be compared across species.
Weaknesses
It would be useful for non-specialists if the authors would introduce each neuropil with some orientation about its function or what kind of input/output it receives, if this is known for other species. Especially those structures that are not described in other arthropods, like the opisthosomal neuropil. Are there implications for neuroanatomical findings in this paper on the understanding of how web-building behaviors are mediated by the brain?
Likewise, where possible, it would be helpful to have some discussion of the implications of certain neurotransmitters/neuropeptides being enriched in different areas. For example, GABA would signal areas of inhibitory connections, such as inhibitory input to mushroom bodies, as described in other arthropods. In the discussion section on relationships between spider and insect midline neuropils, are there similarities in expression patterns between those described here and in insects?
Reviewer #3 (Public review):
Summary:
This is an impressive paper that offers a much-needed 3D standardized brain atlas for the hackled-orb weaving spider Uloborus diversus, an emerging organism of study in neuroethology. The authors used a detailed immunohistological whole-mount staining method that allowed them to localize a wide range of common neurotransmitters and neuropeptides and map them on a common brain atlas. Through this approach, they discovered groups of cells that may form parts of neuropils that had not previously been described, such as the 'tonsillar neuropil', which might be part of a larger insect-like central complex. Further, this work provides unique insights into the previously underappreciated complexity of higher-order neuropils in spiders, particularly the arcuate body, and hints at a potentially important role for the mushroom bodies in vibratory processing for web-building spiders.
Strengths:
To understand brain function, data from many experiments on brain structure must be compiled to serve as a reference and foundation for future work. As demonstrated by the overwhelming success in genetically tractable laboratory animals, 3D standardized brain atlases are invaluable tools - especially as increasing amounts of data are obtained at the gross morphological, synaptic, and genetic levels, and as functional data from electrophysiology and imaging are integrated. Among 'non-model' organisms, such approaches have included global silver staining and confocal microscopy, MRI, and, more recently, micro-computed tomography (X-ray) scans used to image multiple brains and average them into a composite reference. In this study, the authors used synapsin immunoreactivity to generate an averaged spider brain as a scaffold for mapping immunoreactivity to other neuromodulators. Using this framework, they describe many previously known spider brain structures and also identify some previously undescribed regions. They argue that the arcuate body - a midline neuropil thought to have diverged evolutionarily from the insect central complex - shows structural similarities that may support its role in path integration and navigation.
Having diverged from insects such as the fruit fly Drosophila melanogaster over 400 million years ago, spiders are an important group for study - particularly due to their elegant web-building behavior, which is thought to have contributed to their remarkable evolutionary success. How such exquisitely complex behavior is supported by a relatively small brain remains unclear. A rich tradition of spider neuroanatomy emerged in the previous century through the work of comparative zoologists, who used reduced silver and Golgi stains to reveal remarkable detail about gross neuroanatomy. Yet, these techniques cannot uncover the brain's neurochemical landscape, highlighting the need for more modern approaches-such as those employed in the present study.
A key insight from this study involves two prominent higher-order neuropils of the protocerebrum: the arcuate body and the mushroom bodies. The authors show that the arcuate body has a more complex structure and lamination than previously recognized, suggesting it is insect central complex-like and may support functions such as path integration and navigation, which are critical during web building. They also report strong synapsin immunoreactivity in the mushroom bodies and speculate that these structures contribute to vibratory processing during sensory feedback, particularly in the context of web building and prey localization. These findings align with prior work that noted the complex architecture of both neuropils in spiders and their resemblance (and in some cases greater complexity) compared to their insect counterparts. Additionally, the authors describe previously unrecognized neuropils, such as the 'tonsillar neuropil,' whose function remains unknown but may belong to a larger central complex. The diverse patterns of neuromodulator immunoreactivity further suggest that plasticity plays a substantial role in central circuits.
Weaknesses:
My major concern, however, is that some of the authors' neuroanatomical descriptions rely too heavily on inference rather than what is currently resolvable from their immunohistochemistry stains alone.
We would like to thank the reviewers for their time and effort in carefully reading our manuscript and providing helpful feedback, and particularly for their appreciation and realistic understanding of the scope of this study and its context within the existing spider neuroanatomical literature.
Regarding the limitations and potential additions to this study, we believe these to be well-reasoned and are in agreement. We plan to address some of these shortcomings in future publications.
As multiple reviewers remarked, a mapping of the major tracts of the brain would be a welcome addition to understanding the neuroanatomy of U. diversus. This is something which we are actively working on and hope to provide in a forthcoming publication. Given the length of this paper as is, we considered that a treatment of the tracts would be better served as an additional paper. Likewise, mapping of the immunoreactive somata of the currently investigated targets is a component which we would like to describe as part of a separate paper, keeping the focus of the current one on neuropils, in order to leverage our aligned volumes to describe co-expression patterns, which is not as useful for the more widely dispersed somata. Furthermore, while we often see somata through immunostaining, the presence and intensity of the signal is variable among immunoreactive populations. We are finding that these populations are more consistently and comprehensively revealed thru fluorescent in situ hybridization.
We appreciate the desire of the reviewers for further information regarding the connectivity and function of the described neuropils, and where possible we have added additional statements and references. That being said, where this context remains sparse is largely a reflection of the lack of information in the literature. This is particularly the case for functional roles for spider neuropils, especially higher order ones of the protocerebrum, which are essentially unexamined. As summarized in the quite recent update to Foelix’s Spider Neuroanatomy, a functional understanding for protocerebral neuropil is really only available for the visual pathway. Consequently, it is therefore also difficult to speak of the implications for presence or absence of particular signaling elements in these neuropils, if no further information about the circuitry or behavioral correlates are available. Finally, multiple reviewers suggested that it might be worthwhile to explore a comparison of the arcuate body layer innervation to that of the central bodies of insects, of which there is a richer literature. This is an idea which we were also initially attracted to, and have now added some lines to the discussion section. Our position on this is a cautious one, as a series of more recent comparative studies spanning many insect species using the same antibody, reveals a considerable amount of variation in central body layering even within this clade, which has given us pause in interpreting how substantive similarities and differences to the far more distant spiders would be. Still, this is an interesting avenue which merits an eventual comprehensive analysis, one which would certainly benefit from having additional examples from more spider species, in order to not overstate conclusions based on the currently limited neuroanatomical representation.
Given our framing for the impetus to advance neuroanatomical knowledge in orb-web builders, the question of whether the present findings inform the circuitry controlling web-building is one that naturally follows. While we are unable with this dataset alone to define which brain areas mediate web-building - something which would likely be beyond any anatomical dataset lacking complementary functional data – the process of assembling the atlas has revealed structures and defined innervation patterns in previously ambiguous sectors of the spider brain, particularly in the protocerebrum. A simplistic proposal is that such regions, which are more conspicuous by our techniques and in this model species, would be good candidates for further inquiries into web-building circuitry, as their absence or oversight in past work could be attributable to the different behavioral styles of those model species. Regardless, granted that such a hypothesis cannot be readily refuted by the existing neuroanatomical literature, underscores the need to have more finely refined models of the spider brain, to which we hope that we have positively contributed to and are gratified by the reviewer’s enthusiasm for the strengths of this study.
Recommendations for the authors:
Reviewer #1 (Recommendations for the authors):
(1) Brenneis 2022 has done a very nice and comprehensive study focused on the visual system - this might be worth including.
Thank you, we have included this reference on Line 34.
(2) L 29: When talking about "connectivity maps", the emerging connectomes based on EM data could be mentioned.
Additional references have been added, thank you. Line 35.
(3) L 99: Please mention that you are going to describe the brain from ventral to dorsal.
Thank you, we have added a comment to Line 99.
(4) L 13: is found at the posterior.
Thank you, revised.
(5) L 168: How did you pick those two proctolin+ somata, given that there is a lot of additional punctate signal?
Although not visible in this image, if you scroll through the stack there is a neurite which extends from these neurons directly to this area of pronounced immunoreactivity.
(6) Figure 1: Please add the names of the neuropils you go through afterwards.
We have added labels for neuropils which are recognizable externally.
(7) Figure 1 and Figure 5: Please mark the esophagus.
Label has now been added to Figure 1. In Figure 5, the esophagus should not really be visible because these planes are just ventral to its closure.
(8) Figure 5A: I did not see any CCAP signal where the arrow points to; same for 5B (ChAT).
In hindsight, the CCAP point is probably too minor to be worth mentioning, so we have removed it.
The ChAT signal pattern in 5B has been reinforced by adding a dashed circle to show its location as well.
(9) L 249: Could the circular spot also be a tract (many tracts lack synapsin - at least in insects)?
Yes, thank you for pointing this out – the sentence is revised (L274). We are currently further analyzing anti-tubulin volumes and it seem that indeed there are tracts which occupy these synapsin-negative spaces, although interestingly they do not tend to account for the entire space.
(10) L 302: Help me see the "conspicuous" thing.
Brace added to Fig. 8B, note in caption.
(11) L 315: Please first introduce the number of the eyes and how these relate to 1{degree sign} and 2{degree sign} pathway. Are these separate pathways from separate eyes or two relay stations of one visual pathway?
We have expanded the introduction to this section (L336). Yes, these are considered as two separate visual pathways, with a typical segregation of which eyes contribute to which pathway – although there is evidence for species-specific differences in these contributions. In the context of this atlas, we are not currently able to follow which eyes are innervating which pathway.
(12) L 343: It seems that the tonsillar neuropil could be midline spanning (at least this is how I interpret the signal across the midline). Would it make sense to re-formulate from a paired structure to midline-spanning? Would that make it another option for being a central complex homolog?
In the spectrum from totally midline spanning and unpaired (e.g., arcuate body (at least in adults)) to almost fully distinct and paired (e.g., mushroom bodies (although even here there is a midline spanning ‘bridge’)), we view the tonsillar to be more paired due to the oval components, although it does have a midline spanning section, particularly unambiguous just posterior to the oval sections.
Regarding central complex homology, if the suggestion is that the tonsillar with its midline spanning component could represent the entire central complex, then this is a possibility, but it would neglect the highly innervated and layered arcuate body, which we think represent a stronger contender – at least as a component of the central complex. For this reason, we would still be partial to the possibility that the tonsillar is a part of the central complex, but not the entire complex.
(13) L 407: ...and dorsal (..) lobe...
Added the word ‘lobe’ to this sentence (L429).
(14) L 620ff: Maybe mention the role of MBs in learning and memory.
A reference has been added at L661.
(15) L 644: In the context of arcuate body homology with the central body, I was missing a discussion of the neurotransmitters expressed in the respective parts in insects. Would that provide additional arguments?
This is an interesting comparison to explore, and is one that we initially considered making as well. There are certainly commonalities that one could point to, particularly in trying to build the case of whether particular lobes of the arcuate body are similar to the fan-shaped or ellipsoid bodies in insects. Nevertheless, something which has given us pause is studying the more recent comparative works between insect species (Timm et al., 2021, J Comp Neuro, Homberg et al., 2023, J Comp Neuro), which also reveal a fair degree of heterogeneity in expression patterns between species – and this is despite the fact that the neuropils are unambiguously homologous. When comparing to a much more evolutionarily distant organism such as the spider, it becomes less clear which extant species should serve as the best point of comparison, and therefore we fear making specious arguments by focusing on similarities when there are also many differences. We have added some of these comments to the discussion (L699-725).
Throughout the text, I frequently had difficulties in finding the panels right away in the structures mentioned in the text. It would help to number the panels (e.g., 6Ai, Aii, Aii,i etc) and refer to those in the text. Further, all structures mentioned in the text should be labelled with arrows/arrowheads unless they are unequivocally identified in the panel
Thank you for the suggestion. We have adopted the additional numbering scheme for panels, and added additional markers where suggested.
Reviewer #2 (Recommendations for the authors):
(1) L 18: "neurotransmitter" should be pluralized.
Thank you, revised (L18).
(2) L 55: Missing the word "the" before "U. diversus".
Thank you, revised (L57).
(3) L 179: Change synaptic dense to "synapse-dense".
Thank you, revised (L189).
(4) L 570: "present in" would be clearer than "presented on in".
Our intention here was to say that Loesel et al did not show slices from the subesophageal mass for CCAP, so it was ambiguous as to whether it had immunoreactivity there but they simply did not present it, or if it indeed doesn’t show signal in the subesophageal. But agreed, this is awkward phrasing which has been revised (L606-608), thank you.
(5) L 641: It would be worth noting that the upper and lower central bodies are referred to as the fan-shaped and ellipsoid bodies in many insects.
Thank you, this has been added in L694.
(6) L 642: Although cited here regarding insect central body layers, Strausfeld et al. 2006 mainly describe the onychophoran brain and the evolutionary relationship between the onychophoran and chelicerate arcuate bodies. The phylogenetic relationships described here would strengthen the discussion in the section titled "A spider central complex?"
The phylogenetic relationship of onychophorans and chelicerates remains controversial and therefore we find it tricky to use this point to advance the argument in that discussion section, as one could make opposing arguments. The homology of the arcuate body (between chelicerates, onychophorans, and mandibulates) has likewise been argued over, with this Strausfeld et al paper offering one perspective, while others are more permissive (good summary at end of Doeffinger et al., 2010). Our thought was simply to draw attention to grossly similar protocerebral neuropils in examples from distantly related arthropods, without taking a stance, as our data doesn’t really deeply advance one view over the other.
(7) L 701- Noduli have been described in stomatopods (Thoen et al., Front. Behav. Neurosci., 2017).
This is an important addition, thank you – it has been incorporated and cited (L766).
(8) Antisera against DC0 (PKA-C alpha) may distinguish globuli cells from other soma surrounding the mushroom bodies, but this may be accomplished in future studies.
Agreed, this is something we have been interested in, but have not yet acquired the antibody.
Reviewer #3 (Recommendations for the authors):
Overall, this paper is both timely and important. However, it may face some resistance from classically trained arthropod neuroanatomists due to the authors' reliance on immunohistochemistry alone. A method to visualize fiber tracts and neuropil morphology would have been a valuable and grounding complement to the dataset and can be added in future publications. Tract-tracing methods (e.g., dextran injections) would strengthen certain claims about connectivity - particularly those concerning the mushroom bodies. For delineating putative cell populations across regions, fluorescence in situ hybridization for key transcripts would offer convincing evidence, especially in the context of the arcuate body, the tonsillar neuropil, and proposed homologies to the insect central complex.
That said, the dataset remains rich and valuable. Outlined below are a number of issues the authors may wish to address. Most are relatively minor, but a few require further clarification.
(1) Abstract
(a) L 12-14: The authors should frame their work as a novel contribution to our understanding of the spider brain, rather than solely as a tool or stepping stone for future studies. The opening sentences currently undersell the significance of the study.
Thank you for your encourament! We have revised the abstract.
(b) Rather than touting "first of its kind" in the abstract, state what was learned from this.
Thank you, we have revised the abstract.
(c) The abstract does not mention the major results of the study. It should state which brain regions were found. It should list all of the peptides and transmitters that were tested so that they can be discoverable in searches.
Thank you, revised.
(2) Introduction
(a) L 38: There's a more updated reference for Long (2016): Long, S. M. (2021). Variations on a theme: Morphological variation in the secondary eye visual pathway across the order of Araneae. Journal of Comparative Neurology, 529(2), 259-280.
Thank you, this has been updated (L41 and elsewhere).
(b) L 47: While whole-mount imaging offers some benefits, a downside is the need for complete brain dissection from the cuticle, which in spiders likely damages superficial structures (such as the secondary eye pathways).
True – we have added this caveat to the section (L48-51).
(c) L 49-52: If making this claim, more explicit comparisons with non-web building C. saeli in terms of neuropil presence, volume, or density later in the paper would be useful.
We do not have the data on hand to make measured comparisons of C. salei structures, and the neuropils identified in this study are not clearly identifiable in the slices provided in the literature, so would likely require new sample preparations. We’ve removed the reference to proportionality and softened this sentence slightly – we are not trying to make a strong claim, but simply state that this is a possibility.
(3) Results
(a) The authors should state how they accounted for autofluorescence.
While we did not explicitly test for autofluorescence, the long process of establishing a working whole-mount immuno protocol and testing antibodies produced many examples of treated brains which did not show any substantial signal. We have added a note to the methods section (L866).
(b) L 69: There is some controversy in delineating the subesophageal and supraesophageal mass as the two major divisions despite its ubiquity in the literature. It might be safer to delineate the protocerebrum, deutocerebrum, and fused postoral ganglia (including the pedipalp ganglion) instead.
Thank you for this insight, we have modified the section, section headings and Figure 1 to account for this delineation as well. We have chosen to include both ways of describing the synganglion, in order to maintain a parallel with the past literature, and to be further accessible to non-specialist readers. L73-77
(c) L 90: It might be useful to include a justification for the use of these particular neuropeptides.
Thank you, revised. L97-99.
(d) L 106 - 108: It is stated that the innervation pattern of the leg neuropils is generally consistent, but from Figure 2, it seems that there are differences. The density of 5HT, Proctolin, ChAT, and FMRFamide seems to be higher in the posterior legs. AstA seems to have a broader distribution in L1 and is absent in L4.
We would still stand by the generalization that the innervation pattern is fairly similar for each leg. The L1 neuropils tend to be bigger than the posterior legs, which might explain the difference in density. Another important aspect to keep in mind is that not all of the leg neuropils appear at the exact same imaging plane as we move from ventral to dorsal. If you scroll through the synapsin stack (ventral to dorsal), you will see that L2 and L3 appear first, followed shortly by L1, and then L4, and at the dorsal end of the subesophageal they disappear in the opposite order. The observations listed here are true for the single z-plane in Figure 2, but the fact that they don’t appear at the same time seems to mainly account for these differences. For example, if you scroll further ventrally in the AstA volume, you will see a very similar innervation appear in L4 as well, even though it is absent in the Fig. 2 plane. We plan to have these individual volumes available from a repository so that they can be individually examined to better see the signal at all levels. At the moment, the entire repository can be accessed here: https://doi.org/10.35077/ace-moo-far.
(e) Figure 1 and elsewhere: The axes for the posterior and lateral views show Lateral and Medial. It would be more accurate to label them Left and Right. because it does not define the medial-to-lateral axis. The medial direction is correct for only one hemiganglion, and it's the opposite for the contralateral side.
Thank you, revised.
(f) In Figures that show particular sections, it might be helpful to include a plane in the standard brain to illustrate where that section is.
Yes, we agree and it was our original intention. It is something we can attempt to do, but there is not much room in the corners of many of the synapsin panels, making it harder to make the 3D representation big enough to be clear.
(g) Figure 2, 3: Presenting the z-section stack separately in B and C is awkward because it makes it seem that they are unrelated. I think it would be better to display the z160-190 directly above its corresponding z230-260 for each of the exemplars in B and C. Since there's no left-right asymmetry, a hemibrain could be shown for all examples as was done for TH in D. It's not clear why TH was presented differently.
Thank you for this suggestion. We rearranged the figure as described, but ultimately still found the original layout to be preferrable, in part because the labelling becomes too cramped. We hope that the potential confusion of the continuity of the B and C sections will be mitigated by focusing on the z plane labels and overall shape – which should suggest that the planes are not far from each other. We trust that the form of the leg neuropils is recognizable in both B and C synapsin images, and so readers will make the connection.
Regarding TH, this panel is apart from the rest because we were unable to register the TH volume to the standard brain because the variant of the protocol which produced good anti-TH staining conflicted with synapsin, and we could not simultaneously have adequate penetration of the synapsin signal. We did not want to align the TH panel with the others to avoid potential confusion that this was a view from the same z-plane of a registered volume, as the others are. We have added a note to the figure caption.
(h) The locations of the labels should be consistent. The antisera are below the images in Figure 2, above in Figure 3, and to the bottom left in Figure 5. The slices are shown above in Figure 2 and below in Figure 3.
Thank you, this has been revised for better consistency.
(i) It is surprising to me that there is no mention of the neuronal somata visible in Figure 2 and Figure 3. A typical mapping of the brain would map the locations of the neurons, not just the neuropils.
Our first arrangement of this paper described each immunostain individually from ventral to dorsal, including locations of the immunoreactive somata which could be observed. To aid the flow of the paper and leverage the aligned volumes to emphasize co-expression in the function divisions of the brain, we re-formulated to this current layout which is organized around neuropils. Somata locations are tricky to incorporate in this format of the paper which focuses on key z-planes or tight max projections, because the relevant immunoreactive somata are more dispersed throughout the synganglion, not always overlapping in neighboring z-planes. Further, since only a minority of the antisera we used can reveal traceable projections from the supplying somata in the whole-mount preparation, we would be quite limited in the degree to which we could integrate the specific somata mapping with expression patterns in the neuropil. Finally, compared to immuno, which can be variable in staining intensity between somata for the same target, we find that FISH reveals these locations more clearly and comprehensively – so while we agree that this mapping would also be useful for the atlas, we would like to better provide this information in a future publication using whole-mount FISH.
(j) L 139: There is a reference to a "brace" in Figure 3B, which does not seem to exist. There's one in Figure 3C.
There is a smaller brace near the bottom of the TDC2 panel in Fig. 3B.
(k) L 151 should be "3D".
Thank you, revised (L160).
(l) Figure 4C: It is not mentioned in the legend that the bottom inset is Proctolin without synapsin.
Thank you, revised (L1213).
(m) L 199: Are the authors sure this subdivision is solely on the anterior-posterior axis? Could it also be dorsal ventral? (i.e., could this be an artifact of the protocerebrum and deutocerebrum?)
Yes, this division can be appreciated to extend somewhat in the dorsal-ventral axis and it is possible that this is the protocerebrum emerging after the deutocerebrum, although this area is largely dorsal to the obvious part of the deutocerebrum. In the horizontal planes there appears to be a boundary line which we use for this subdivision in order to assist in better describing features within this generally ventral part of the protocerebrum – referred to as “stalk” because it is thinner before the protocerebrum expands in size, dorsally. Our intention was more organizational, and as stated in the text, this area is likely heterogenous and we are not suggesting that it has a unified function, so being a visual artifact would not be excluded.
(n) L 249: Could it also indicate large tracts projecting elsewhere?
Yes, definitely, we have evidence that part of the space is occupied by tracts. Revised, thank you (L262).
(o) L 281: Several investigators, including Long (2021,) noted very large and robust mushroom bodies of Nephila.
Thank you – the point is well taken that there are examples of orb-web builders that do have appreciable mushroom bodies. We have added a note in this section (L295), giving the examples of Deinopis spinosa and Argiope trifasciata (Figure 4.20 and 4.22 in Long, 2016).
It looks like these species make the point better than Nephila, as Long lists the mushroom body percentage of total protocerebral volume for D. spinosa as 4.18%, for A. trifasciata as 2.38%, but doesn’t give a percentage for Nephila clavipes (Figure 4.24) and only labels the mushroom bodies structures as “possible” in the figure.
In Long (2021), Nephilidae is described as follows: “In Nephilidae, I found what could be greatly reduced medullae at the caudal end of the laminae, as well as a structure that has many physical hallmarks of reduced mushroom bodies”
(p) L 324: If the authors were able to stain for histamine or supplement this work with a different dissection technique for the dorsal structures, the visual pathways might have been apparent, which seems like a very important set of neuropils to include in a complete brain atlas.
Yes, for this reason histamine has been an interesting target which we have attempted to visualize, but unfortunately have not yet been able to successfully stain for in U. diversus. An additional complication is that the antibodies we have seen call for glutaraldehyde fixation, which may make them incompatible with our approach to producing robust synapsin staining throughout the brain.
We agree that the lack of the complete visual pathway is a substantial weakness of our preparation, and should be amended in future work, but this will likely require developing a modified approach in order to preserve these delicate structures in U. diversus.
(q) L 331: Is this bulbous shape neuropil, or just the remains of neuropil that were not fully torn away during dissection?
This certainly is a severed part of the primary pathway, although it seems more likely that the bulbous shape is indicative of a neuropil form, rather than just being a happenstance shape that occurred during the breakage. We have examples where the same bulbous shape appears on both sides, and in different brains. It is possible that this may be the principal eye lamina – although we did not see co-staining with expected markers in examples where it did appear, so cannot be sure.
(r) L 354: Is tyraminergic co-staining with the protocerebral bridge enough evidence to speculate that inputs are being supplied?
We agree that this is not compelling, and have removed the statement.
(s) L 372: This whole structure appears to be a previously described structure in spiders, the 'protocerebral commissure'.
We are reasonably sure that what we are calling the PCB is a distinct structure from the protocerebral bridge (PCC). In Babu and Barth’s (1984) horizontal slice (Fig. 11b), you can see the protocerebral commissure immediately adjacent to the mushroom body bridge. It is found similarly located in other species, as can be seen in the supplementary 3D files provided by Steinhoff et al., (2024).
While not visible with synapsin in U. diversus, we likewise can make out a commissure in this area in close proximity to the mushroom body bridge using tubulin staining. What we are calling the protocerebral bridge is a structure which is much more dorsal to the protocerebral commissure, not appearing in the same planes as the MB bridge.
(t) L 377: Do you have an intuition why the tonsillar neuropil and the protocerebral bridge would show limited immunoreactivity, while the arcuate body's is quite extensive?
This is an interesting question. Given the degree of interconnection and the fact that multiple classes of neurons in insects will innervate both central body as well as PCB or noduli, perhaps it would be expected that expression in tonsillar and protocerebral bridge should be commensurate to the innervation by that particular neurotransmitter expressing population in the arcuate body. Apart from the fact that the arcuate body is just bigger, perhaps this points to a great role of the arcuate body for integration, whereas the tonsillar and PCB may engage in more particular processing, or be limited to certain sensory modalities.
Interestingly, it seems that this pattern of more limited immunoreactivity in the PCB and noduli compared with the central bodies (fan-shaped/ellipsoid) also appears in insects (Kahsai et al., 2010, J Comp Neuro, Timm et al., 2021, J Comp Neuro, Homberg et al., 2023, J Comp Neuro) – particularly, with almost every target having at least some layering in the fan-shaped body (Kahsai et al., 2010, J Comp Neuro). For example, serotoninergic innervation is fairly consistently seen in the upper and lower central bodies across insects, but its presence in the PCB or noduli is more variable – appearing in one or the other in a species-dependent manner (Homberg et al., 2023, J Comp Neuro).
(4) Discussion
(a) L 556: But if confocal images from slices are aligned, is the 3D shape not preserved?
Yes, fair enough – the point we wanted to make was that there is still a limitation in z resolution depending on the thickness of the slices used, which could obscure structures, but perhaps this is too minor of a comment.
(b) L 597: This is a very interesting result. I agree it's likely to do with the processing of mechanosensory information relevant to web activities, and the mushroom body seems like the perfect candidate for this.
(c) L 638: Worth noting that neuropil volume vs density of synapses might play a role in this, as the literature is currently a bit ambiguous with regards to the former.
Thank you, noted (L689).
(d) L 651: The latter seems far more plausible.
Agreed, though the presence of mushroom bodies appears to be variable in spiders, so we didn’t want to take a strong stance, here.
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the original reviews.
Public Reviews:
Reviewer #1 (Public review)
Major:
(1) In line 76, the authors make a very powerful statement: 'σRNN simulation achieves higher similarity with unseen recorded trials before perturbation, but lower than the bioRNN on perturbed trials.' I couldn't find a figure showing this. This might be buried somewhere and, in my opinion, deserves some spotlight - maybe a figure or even inclusion in the abstract.
We agree with the reviewer that these results are important. The failure of σRNN on perturbed data could be inferred from the former Figures 1E, 2C-E, and 3D. Following the reviewers' comments, we have tried to make this the most prominent message of Figure 1, in particular with the addition of the new panel E. We also moved Table 1 from the Supplementary to the main text to highlight this quantitatively.
(2) It's mentioned in the introduction (line 84) and elsewhere (e.g., line 259) that spiking has some advantage, but I don't see any figure supporting this claim. In fact, spiking seems not to matter (Figure 2C, E). Please clarify how spiking improves performance, and if it does not, acknowledge that. Relatedly, in line 246, the authors state that 'spiking is a better metric but not significant' when discussing simulations. Either remove this statement and assume spiking is not relevant, or increase the number of simulations.
We could not find the exact quote from the reviewer, and we believe that he intended to quote “spiking is better on all metrics, but without significant margins”. Indeed, spiking did not improve the fit significantly on perturbed trials, this is particularly true in comparison with the benefits of Dale’s law and local inhibition. As suggested by the reviewer, we rephrased the sentence from this quote and more generally the corresponding paragraphs in the intro (lines 83-87) and in the results (lines 245-271). Our corrections in the results sections are also intended to address the minor point (4) raised by the same reviewer.
(3) The authors prefer the metric of predicting hits over MSE, especially when looking at real data (Figure 3). I would bring the supplementary results into the main figures, as both metrics are very nicely complementary. Relatedly, why not add Pearson correlation or R2, and not just focus on MSE Loss?
In Figure 3 for the in-vivo data, we do not have simultaneous electrophysiological recordings and optogenetic stimulation in this dataset. The two are performed on different recording sessions. Therefore, we can only compare the effect of optogenetics on the behavior, and we cannot compute Pearson correlation or R2 of the perturbed network activity. To avoid ambiguity, we wrote “For the sessions of the in vivo dataset with optogenetic perturbation that we considered, only the behavior of an animal is recorded” on line 294.
(4) I really like the 'forward-looking' experiment in closed loop! But I felt that the relevance of micro perturbations is very unclear in the intro and results. This could be better motivated: why should an experimentalist care about this forward-looking experiment? Why exactly do we care about micro perturbation (e.g., in contrast to non-micro perturbation)? Relatedly, I would try to explain this in the intro without resorting to technical jargon like 'gradients'.
As suggested, we updated the last paragraph of the introduction (lines 88 - 95) to give better motivation for why algorithmically targeted acute spatio-temporal perturbations can be important to dissect the function of neural circuits. We also added citations to recent studies with targeted in vivo optogenetic stimulation. As far as we know the existing previous work targeted network stimulation mostly using linear models, while we used non-linear RNNs and their gradients.
Minor:
(1) In the intro, the authors refer to 'the field' twice. Personally, I find this term odd. I would opt for something like 'in neuroscience'.
We implemented the suggested change: l.27 and l.30
(2) Line 45: When referring to previous work using data-constrained RNN models, Valente et al. is missing (though it is well cited later when discussing regularization through low-rank constraints)
We added the citation: l.45
(3) Line 11: Method should be methods (missing an 's').
We fixed the typo.
(4) In line 250, starting with 'So far', is a strange choice of presentation order. After interpreting the results for other biological ingredients, the authors introduce a new one. I would first introduce all ingredients and then interpret. It's telling that the authors jump back to 2B after discussing 2C.
We restructured the last two paragraphs of section 2.1, and we hope that the presentation order is now more logical.
(5) The black dots in Figure 3E are not explained, or at least I couldn't find an explanation.
We added an explanation in the caption of Figure 3E.
Reviewer #2 (Public review):
(1) Some aspects of the methods are unclear. For comparisons between recurrent networks trained from randomly initialized weights, I would expect that many initializations were made for each model variant to be compared, and that the performance characteristics are constructed by aggregating over networks trained from multiple random initializations. I could not tell from the methods whether this was done or how many models were aggregated.
The expectation of the reviewer is correct, we trained multiple models with different random seeds (affecting both the weight initialization and the noise of our model) for each variant and aggregated the results. We have now clarified this in Methods 4.6. lines 658-662.
(2) It is possible that including perturbation trials in the training sets would improve model performance across conditions, including held-out (untrained) perturbations (for instance, to units that had not been perturbed during training). It could be noted that if perturbations are available, their use may alleviate some of the design decisions that are evaluated here.
In general, we agree with the reviewer that including perturbation trials in the training set would likely improve model performance across conditions. One practical limitation explaining partially why we did not do it with our dataset is the small quantity of perturbed trials for each targeted cortical area: the number of trials with light perturbations is too scarce to robustly train and test our models.
More profoundly, to test hard generalizations to perturbations (aka perturbation testing), it will always be necessary that the perturbations are not trivially represented in the training data. Including perturbation trials during training would compromise our main finding: some biological model constraints improve the generalization to perturbation. To test this claim, it was necessary to keep the perturbations out of the training data.
We agree that including all available data of perturbed and non-perturbed recordings would be useful to build the best generalist predictive system. It could help, for instance, for closed-loop circuit control as we studied in Figure 5. Yet, there too, it will be important for the scientific validation process to always keep some causal perturbations of interest out of the training set. This is necessary to fairly measure the real generalization capability of any model. Importantly, this is why we think out-of-distribution “perturbation testing” is likely to have a recurring impact in the years to come, even beyond the case of optogenetic inactivation studied in detail in our paper.
Recommendation for the authors:
Reviewer #1 (Recommendation for the authors):
The code is not very easy to follow. I know this is a lot to ask, but maybe make clear where the code is to train the different models, which I think is a great contribution of this work? I predict that many readers will want to use the code and so this will improve the impact of this work.
We updated the code to make it easier to train a model from scratch.
Reviewer #2 (Recommendation for the authors):
The figures are really tough to read. Some of that small font should be sized up, and it's tough to tell in the posted paper what's happening in Figure 2B.
We updated Figures 1 and 2 significantly, in part to increase their readability. We also implemented the "Superficialities" suggestions.
-
-
courses.ecu.edu.au courses.ecu.edu.au
-
Download Your Action Plan Toolkit
The Feedback Action Plan Template is a strong and practical resource, and it’s an ideal place to make the three movements of Engagement (⇄E) explicit. Rather than renaming the template, we suggest lightly structuring it around Reflection, Inquiry, and Action — using labels or prompts to help learners see Engagement as a process they practise over time. Much of this is already present; the main enhancement would be distinguishing an initial reflection on how feedback lands (emotionally and cognitively) from later reflection on capability development, and clearly positioning SAG insights as part of the Inquiry move.
Suggested enhancements to the Feedback Action Plan Template 1. Keep the title “Feedback Action Plan Template” The title is clear and learner-friendly. Rather than renaming it, the conceptual work can be done through how the template is structured and framed. 2. Add a brief framing line at the top to connect the template to ⇄E For example: “This Action Plan helps you practise the Engagement (⇄E) part of the SAG⇄E Insights for Learning framework by guiding you through three moves: Reflection, Inquiry, and Action.” 3. Make the three ⇄E movements explicit through light section labelling or prompts This helps learners see Engagement as a process they practise, not just a single step. 4. Surface an initial Reflection move (before action planning) Add a short reflection prompt that invites learners to notice how feedback lands emotionally and cognitively, for example: • What stood out to you in this feedback? • How did it make you feel or think differently about your work or learning? This normalises reflection and supports learning from challenge or mistakes. 5. Position SAG insights as part of the Inquiry move Reframe the existing “SAG⇄E Insights” section as Inquiry, e.g.: Which Successes, Adjustments, or Growth insights matter most right now, and why? This reinforces that SAG insights are inputs to learner sense-making, not endpoints. 6. Retain the Action section with minimal change The current focus on specific, achievable steps is strong. Optional prompts could reinforce time-bounded action (e.g. “over the next week or two”). 7. Differentiate reflection on ‘how feedback landed’ from reflection on development over time The existing “Reflection on Capability Development” section works well as a later reflection, focused on noticing learning, growth, or changes in confidence after acting on feedback. 8. Keep Support Resources and Portfolio Annotation as they are, with minor connective language if helpful These sections already align well with ⇄E and portfolio learning; small wording tweaks could simply reinforce their role in supporting the Engagement process.
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the original reviews.
Public Reviews:
Reviewer #1 (Public review):
Summary:
This work by Al-Jezani et al. focused on characterizing clonally derived MSC populations from the synovium of normal and osteoarthritis (OA) patients. This included characterizing the cell surface marker expression in situ (at time of isolation), as well as after in vitro expansion. The group also tried to correlate marker expression with trilineage differential potential. They also tested the ability of the different subpopulations for their efficacy in repairing cartilage in a rat model of OA. The main finding of the study is that CD47hi MSCs may have a greater capacity to repair cartilage than CD47lo MSCs, suggesting that CD47 may be a novel marker of human MSCs that have enhanced chondrogenic potential.
Strengths:
Studies on cell characterization of the different clonal populations isolated indicate that the MSC are heterogenous and traditional cell surface markers for MSCs do not accurately predict the differentiation potential of MSCs. While this has been previously established in the field of MSC therapy, the authors did attempt to characterize clones derived from single cells, as well as evaluate the marker profile at the time of isolation. While the outcome of heterogeneity is not surprising, the methods used to isolate and characterize the cells were well developed. The interesting finding of the study is the identification of CD47 as a potential MSC marker that could be related to chondrogenic potential. The authors suggest that MSCs with high CD47 repaired cartilage more effectively than MSC with low CD47 in a rat OA model.
Weaknesses:
While the identification of CD47 as a novel MSC marker could be important to the field of cell therapy and cartilage regeneration, there was a lack of robust data to support the correlation of CD47 expression to chondrogenesis. The authors indicated that the proteomics suggested that the MSC subtype expressed significantly more CD47 than the non-MSC subtype. However, it was difficult to appreciate where this was shown. It would be helpful to clearly identify where in the figure this is shown, especially since it is the key result of the study. The authors were able to isolate CD47hi and CD47 low cells. While this is exciting, it was unclear how many cells could be isolated and whether they needed to be expanded before being used in vivo. Additional details for the CD47 studies would have strengthened the paper. Furthermore, the CD47hi cells were not thoroughly characterized in vitro, particularly for in vitro chondrogenesis. More importantly, the in vivo study where the CD47hi and CD47lo MSCs were injected into a rat model of OA lacked experimental details regarding how many cells were injected and how they were labeled. No representative histology was presented and there did not seem to be a statistically significant difference between the OARSI score of the saline injected and MSC injected groups. The repair tissue was stained for Sox9 expression, which is an important marker of chondrogenesis but does not show production of cartilage. Expression of Collagen Type II would be needed to more robustly claim that CD47 is a marker of MSCs with enhanced repair potential.
Reviewer #2 (Public review):
Summary:
This is a compelling study that systematically characterized and identified clonal MSC populations derived from normal and osteoarthritis human synovium. There is immense growth in the focus on synovial-derived progenitors in the context of both disease mechanisms and potential treatment approaches, and the authors sought to understand the regenerative potential of synovial-derived MSCs.
Strengths:
This study has multiple strengths. MSC cultures were established from an impressive number of human subjects, and rigorous cell surface protein analyses were conducted, at both pre-culture and post-culture timepoints. In vivo experiments using a rat DMM model showed beneficial therapeutic effects of MSCs vs non-MSCs, with compelling data demonstrating that only "real" MSC clones incorporate into cartilage repair tissue and express Prg4. Proteomics analysis was performed to characterize non-MSC vs MSC cultures, and high CD47 expression was identified as a marker for MSC. Injection of CD47-Hi vs CD47-Low cells in the same rat DMM model also demonstrated beneficial effects, albeit only based on histology. A major strength of these studies is the direct translational opportunity for novel MSC-based therapeutic interventions, with high potential for a "personalized medicine" approach.
Weaknesses:
Weaknesses of this study include the rather cursory assessment of the OA phenotype in the rat model, confined entirely to histology (i.e. no microCT, no pain/behavioral assessments, no molecular readouts). It is somewhat unclear how the authors converged on CD47 vs the other factors identified in the proteomics screen, and additional information is needed to understand whether true MSCs only engraft in articular cartilage or also in ectopic cartilage (in the context of osteophyte/chondrophyte formation). Some additional discussion and potential follow-up analyses focused on other cell surface markers recently described to identify synovial progenitors is also warranted. A conceptual weakness is the lack of discussion or consideration of the multiple recent studies demonstrating that DPP4+ PI16+ CD34+ stromal cells (i.e. the "universal fibroblasts") act as progenitors in all mesenchymal tissues, and their involvement in the joint is actively being investigated. Thus, it seems important to understand how the MSCs of the present study are related to these DPP4+ progenitors. Despite these areas for improvement, this is a strong paper with a high degree of rigor, and the results are compelling, timely, and important.
Overall, the authors achieved their aims, and the results support not just the therapeutic value of clonally-isolated synovial MSCs but also the immense heterogeneity in stromal cell populations (containing true MSCs and non-MSCs) that must be investigated further. Of note, the authors employed the ISCT criteria to characterize MSCs, with mixed results in pre-culture and post-culture assessments. This work is likely to have a longterm impact on methodologies used to culture and study MSCs, in addition to advancing the field's knowledge about how synovial-derived progenitors contribute to cartilage repair in vivo.
Recommendations for the authors:
Reviewer #1 (Recommendations for the authors):
In all figures, it would be beneficial to report the sample number used for the data reported. It is difficult to appreciate the statistical analysis without that information.
Understood, the sample number and replicates have been added to each figure legend.
Please check that Table S7 is part of the manuscript. It could not be found.
It was added as an additional excel file since it was too large to fit in the word document.
Lines 377-379 (Figure 2E): the authors write that rats receiving MSCs had a significantly lower OARSI and Krenn score vs. rats injected with non-MSCs. However, none of the bars indicating statistical significance run between these two groups. Please verify the text and figure.
This has been corrected
The details surrounding the labeling of the cells with tdTomato were not presented in the methods.
This has been added to the methods
The fluorescent antibodies used should be listed and more details provided in the methods rather than a general statement that fluorescent antibodies were used.
Our apologies, the clones and companies have been added.
Additional information on the CD47 experiments (# cells, # animals) would have strengthened the study.
This has been added to the methods and figure legend.
Reviewer #2 (Recommendations for the authors):
My comments span minor corrections, requests for additional analyses, some suggestions for additional experiments, and requests for additional discussion of recent important studies.
Introduction:
The introduction is thorough and well-written. I recommend a brief discussion about the emerging evidence demonstrating that DPP4+ PI16+ CD34+ synovial cells, i.e. the "universal fibroblasts", act as stromal progenitors in development, homeostasis, and disease. Relevant osteoarthritis-related papers encompass human and mouse studies (PMIDs: 39375009, 38266107, 38477740, 36175067, 36414376).
This has been added.
Relatedly, as DPP4 is CD26 and therefore useful as a cell-surface antigen for flow cytometry, sorting, etc, it would be interesting to understand the relationship and similarities between the CD47-High cells identified in this study and the DPP4/PI16+ cells previously described. Do they overlap in phenotype/identity?
We have added a new flow cytometry figure for address this question.
Results:
Note type-o on Line 311: "preformed" instead of "performed". Line 313 "prolife" instead of "profile"
Thank you for catching these.
The identified convergence of the cell surface marker profile of all normal and OA clones in culture is a highly intriguing result. Do the authors have stored aliquots of these cells to demonstrate whether this would also occur in soft substrate, i.e. low stiffness culture conditions? This could be done with standard dishes coated with bulk collagen or with commercially available low-stiffness dishes (1 kPa). This is relevant to multiple studies demonstrating the induction of a myofibroblast-like phenotype by stromal cells cultured on high-stiffness plastic or glass. This is also the experiment where assessment of DPP4/CD26 could be added, if possible.
While we agree it would be interesting to determine the mechanism by which the cells phenotypes converge, we would argue that it is outside of the scope of the current manuscript. We have instead added a sentence to the discussion.
Line 353 regarding the use of CD68 as a negative gate: can the authors pleasecomment on why they employed CD68 here and not CD45? While monocytes/macs/myeloid cells are the most abundant immune cells in synovium, CD45 would more comprehensively exclude all immune cells.
That is a fair point, and we really don’t have any reason to have picked CD68 over CD45. In our opinion either would be a fair negative marker to use based on the literature.
Fig 2, minor suggestion: consider adding "MSC vs non-MSC" on the experimental schematic to more comprehensively summarize the experiment.
This has been modified
Fig 2E should show all individual datapoints, not just bar graphs.
This has been modified
Fig 2: Given the significant reduction in Krenn score in DMM-MSC injected knees compared to DMM-saline knees, Fig 2 should also show representative images of the synovial phenotype to demonstrate which aspects of synovial pathology were mitigated. Was the effect related to lining hyperplasia, subsynovial infiltrate, fibrosis, etc? Similarly, can the authors narrate which aspects of the OARSI score drove the treatment effect (proteoglycans vs structure vs osteophytes, etc).
We have added a new sup figure breaking down the Krenn score as well as higher magnification images of representative synovium.
Fig 2: In the absence of microCT imaging, can the authors quantify subchondral bone morphometrics using multiple histological sections? The tibial subchondral bone in Fig 2D appears protected from sclerosis/thickening.
Unfortunately, this is beyond what are able to add to the manuscript.
The Fig 3 results are highly compelling and interesting. Congratulations.
Thank you very much.
Fig 4A: the cell highlighted in the high-mag zoom box in Fig 4A appears to be localized within the joint capsule or patellar tendon (it is unclear which anatomic region this image represents). The highly aligned nature of the tissue and cells along a fibrillar geometry indicates that this is not synovium. The interface between synovium and the tissue in question can be clearly observed in this image. Please choose an image more representative of synovium.
We completely agree with the reviewers assessment. However, it is the synovium that overlays this tissue (Fig 4A arrow). We are simply showing that there were very few MSCs that took up residence in the synovium or the adjacent tissues.
Fig 4C and F: please show individual data points.
This has been added
Fig 5D: I see DPP4 and ITGA5 were also hits in the proteomics analysis, which is intriguing. Besides my comments/suggestions regarding DPP4 above, please note this recent paper identifying a ITGA5+ synovial fibroblast subset that orchestrates pathological crosstalk with lymphocytes in RA, PMID: 39486872
Thank you for the information. We have added the reference in the results section.
Fig 5B-D: How did the authors converge on CD47 as the target for follow-up study? It does not appear to be a differentially-expressed protein based on the Volcano plot in Fig 5B, and it's unclear why it is a more important factor than any of the other proteins shown in the network diagram in Fig 5D, e.g. CTSL, ITGA5, DPP4. Can the authors add a quantitative plot supporting their statement "the MSC sub-type expressed significantly more CD47 than the non-MSCs" on Line 458?
We have re-written this line. It was incorrect to discuss amount of CD47. That was shown later with the flow analysis.
Fig 6D: Please show individual data points and also representative histology images to demonstrate the nature of the phenotypic effect.
This has been added.
Fig 6E-F: In what anatomic region are these images? Please add anatomic markers to clarify the location and allow the reader to interpret whether this is articular cartilage or ectopic cartilage
We have redone the figure to show the area as requested.
Relevant to this, do the authors observe this type of cellular engraftment in ectopic cartilage/osteophytes or only in articular cartilage? Understanding the contribution of these cells to the formation/remodeling of various cartilage types in the context of OA is a critical aspect of this line of investigation.
We didn’t see any contribution of these cells to ectopic cartilage formation and are actively working on a follow up study discussing this point specifically.
Discussion:
Besides my comments regarding DPP4 and ITGA5 above, the authors may also consider discussing PMID: 37681409 (JCI Insight 2023), which demonstrates that adult Prg4+ progenitors derived from synovium contribute to articular cartilage repair in vivo.
We agree that there are numerous markers we could look at in future studies and that other people in the field are actively studying.
-
-
human.libretexts.org human.libretexts.org
-
Logos is most easily defined as the logical appeal of an argument. Say that you are writing a paper on COVID-19 and you say "COVID-19 is just like the flu, so we should take the same measures as the flu." This statement is illogical because the virus itself, it's characteristics, and the overall situation is not like that of the flu. The statement has an illogical comparison. The COVID-19 virus is in a different family of viruses (corona viruses) than are the various influenza viruses, such as H1N1. COVID-19 displays a wide variety of symptoms (or no symptoms) and is much more contagious precisely because it can be transmitted without any symptoms. In addition, we have immunizations against the flu virus, which we do not yet have for the COVID-19 virus.
Logos focuses on logic, facts, and reasoning. Using evidence and clear explanations helps make an argument more convincing and reasonable to the audience.
Tags
Annotators
URL
-
-
www.biorxiv.org www.biorxiv.org
-
Note: This preprint has been reviewed by subject experts for Review Commons. Content has not been altered except for formatting.
Learn more at Review Commons
Referee #1
Evidence, reproducibility and clarity
The authors present an investigation into the surprising effectiveness of the negative-binomial distribution in modelling transcript counts in single cell RNA sequencing experiments. With experimentally motivated ground-truth models that incorporate transcriptional bursting, they show that when transcription activity is large compared to degradation these distributions coincide. With a novel model selection metric, they indicate the regions of parameter space in which the negative-binomial model is a good approximation to the underlying true model. With this procedure, they also indicate that transcriptional burst parameters are unlikely to be reconstructed by an effective negative-binomial function, but that nevertheless, relative rankings between genes can be identified robustly.
I would like to commend the authors on an interesting and fairly comprehensive investigation on a topic of considerable importance in the interpretation of single cell RNA sequencing experiments, and on a well written paper. I have no major comments on issues that affect the conclusions of the paper, although I have a few minor suggestions that might aid reader's understanding of the results and their applicability.
General
It would be nice to have a comparison with some real data for the burst frequency and size, just to indicate to the reader how important these regions are compared to what might be measured. For example, if most genes are outside of the region that does not accommodate the NB distribution, then the conclusion is quite different than if most real counts are unlikely to accommodated by the NB.
Inter-cellular variability of transcription dynamics is quite a significant point of interest, so it would be good to have stated earlier that this is not considered, with the mitigation that is noted later. This is particularly important given that in the introduction, the cases mentioned seem to imply that an NB distribution would be more likely with higher inter-cellular variability.
Introduction
It would be nice to have a bit more detail here, for example on what UMIs are, and what the parameters of the NB distribution represent in general.
For smFISH, I would have thought that the more simple explanation is that the NB is often the simplest distribution with some overdispersion that fits the data, and the parameters don't necessarily need to be biologically interpretable?
It's noted later that the capture probability of modern RNASeq protocols can be ~0.3, which doesn't seem very different compared to 0.7-0.9 of smFISH, so some context here would be good.
Results
Eq 1: I don't think you lose anything by giving the Pochammer symbol and Kummer confluent geometric function explicitly here, and it would make it it a lot easier to read. That said, this equation also seems to come out of nowhere, so a reference would be nice.
I think the moment matching is reasonably convincing, but it might require a little more explicit motivation for a more general audience.
Thm. 1: Do these converge at similar rates, and if not, does that have any implications for the interpretation of the comparisons (as these are evaluated with specific values)? This might be worth a short comment.
Fig 3. In the description for this in the text, it would be nice to have an expression of the KL divergence (and what order the arguments are in), for anyone unfamiliar.
The discussion of the aeBIC seems a bit circuitous. A reasonable prior intention might be to average (or apply a voting function) to individual BIC values, rather than the aeBIC constructed here. And in fact the text goes on to note after the description that this is a good estimate of the expectation of the BIC after all, with some computational advantages. So it might be better to have a more straightforward presentation where this is proposed as an approximation to the expectation of the BIC in the first place.
Section 2.4: The intro to this section could do with a bit more background of the capture, PCR, sequencing, etc, stages, and what exactly the data generated here represents. Otherwise the discussion of zero inflation and UMIs is a little confusing.
It would also be nice to have a comment here on the effect of sequencing depth, or similar (compared to capture probability), even if this wouldn't change the interpretation.
Significance
The paper provides novel arguments towards the support of the negative-binomial distribution in describing single cell RNA sequencing data, with particular relevance to transcriptional bursting observed in numerous datasets. The paper follows from some notable prior work in the field, and integrates these into a more consistent description, particularly in relation to newer techniques such as UMIs.
The ubiquity of the negative-binomial distribution means that these arguments will be of relevance to those that perform theoretical or statistical modelling of single cell RNA sequencing data, and theoretically justifies many widely held assumptions. However, the paper does not make any reference to specific reference datasets or commonly observed values, so where in the parameter space data likely lies would still need to be evaluated on a case-by-case basis.
My expertise is in mathematical modelling and statistics, with some experience of the analysis of single cell RNA sequencing data.
-
-
-
Spionage muss nicht mehr (so) teuer sein Der Fund zeigt auch einen neuen Trend bei staatlicher Überwachung. Bisher setzten Staaten oft auf spezialisierte Spionagesoftware. Das bekannteste Beispiel ist die Anwendung Pegasus, mit der zum Beispiel das Umfeld des ermordeten Journalisten Jamal Khashoggi ausspioniert worden sein soll. Diese Art Software lauert auf Sicherheitslücken in Smartphone-Betriebssystemen wie Android und iOS und nutzt sie dann aus. Das geht auch aus der Ferne, zum Beispiel über Textnachrichten mit Schadcode. Doch diese Art von Spionagesoftware ist teuer. Deshalb nutzen Regime heute verstärkt die Möglichkeit, die ein physischer Zugriff auf das Gerät bietet. Soll heißen: Sie nehmen es an sich und spielen gezielt Spionagesoftware drauf. Zuletzt hatte die Nichtregierungsorganisation Amnesty International eine Spionagesoftware des serbischen Geheimdienstes enttarnt, mit der Journalistinnen und Aktivisten überwacht wurden. Sicherheitsforscher fanden ähnliche Ansätze russischer und chinesischer Behörden, die sowohl Oppositionelle als auch reisende Geschäftsleute überwachen. In allen Fällen hatten die Behörden Verhöre oder andere Gelegenheiten genutzt, um die Geräte zu manipulieren.
Not a 'new trend' just what was always done. Physical access to devices always preferred when it's targeted surveillance. Weird piece of journalism, makes me think that author is new to this topic. Same being said in comments btw
-
-
www.biorxiv.org www.biorxiv.org
-
Reviewer #2 (Public review):
Summary:
This study uses dental traits of a large sample of Chinese mammals to track evolutionary patterns through the Paleocene. It presents and argues for a 'brawn before bite' hypothesis - mammals increased in body size disparity before evolving more specialized or adapted dentitions. The study makes use of an impressive array of analyses, including dental topographic, finite element, and integration analyses, which help to provide a unique insight into mammalian evolutionary patterns.
Strengths:
This paper helps to fill in a major gap in our knowledge of Paleocene mammal patterns in Asia, which is especially important because of the diversification of placentals at that time. The total sample of teeth is impressive and required considerable effort for scanning and analyzing. And there is a wealth of results for DTA, FEA, and integration analyses. Further, some of the results are especially interesting, such as the novel 'brawn before bite' hypothesis and the possible link between shifts in dental traits and arid environments in the Late Paleocene. Overall, I enjoyed reading the paper, and I think the results will be of interest to a broad audience.
Weaknesses:
I have four major concerns with the study, especially related to the sampling of teeth and taxa, that I discuss in more detail below. Due to these issues, I believe that the study is incomplete in its support of the 'brawn before bite' hypothesis. Although my concerns are significant, many of them can be addressed with some simple updates/revisions to analyses or text, and I try to provide constructive advice throughout my review.
(1) If I understand correctly, teeth of different tooth positions (e.g., premolars and molars), and those from the same specimen, are lumped into the same analyses. And unless I missed it, no justification is given for these methodological choices (besides testing for differences in proportions of tooth positions per time bin; L902). I think this creates some major statistical concerns. For example, DTA values for premolars and molars aren't directly comparable (I don't think?) because they have different functions (e.g., greater grinding function for molars). My recommendation is to perform different disparity-through-time analyses for each tooth position, assuming the sample sizes are big enough per time bin. Or, if the authors maintain their current methods/results, they should provide justification in the main text for that choice.
Also, I think lumping teeth from the same specimen into your analyses creates a major statistical concern because the observations aren't independent. In other words, the teeth of the same individual should have relatively similar DTA values, which can greatly bias your results. This is essentially the same issue as phylogenetic non-independence, but taken to a much greater extreme.
It seems like it'd be much more appropriate to perform specimen-level analyses (e.g., Wilson 2013) or species-level analyses (e.g., Grossnickle & Newham 2016) and report those results in the main text. If the authors believe that their methods are justified, then they should explain this in the text.
(2) Maybe I misunderstood, but it sounds like the sampling is almost exclusively clades that are primarily herbivorous/omnivorous (Pantodonta, Arctostylopida, Anagalida, and maybe Tillodonta), which means that the full ecomorphological diversity of the time bins is not being sampled (e.g., insectivores aren't fully sampled). Similarly, the authors say that they "focused sampling" on those major clades and "Additional data were collected on other clades ... opportunistically" (L628). If they favored sampling of specific clades, then doesn't that also bias their results?
If the study is primarily focused on a few herbivorous clades, then the Introduction should be reframed to reflect this. You could explain that you're specifically tracking herbivore patterns after the K-Pg.
(3) There are a lot of topics lacking background information, which makes the paper challenging to read for non-experts. Maybe the authors are hindered by a short word limit. But if they can expand their main text, then I strongly recommend the following:
(a) The authors should discuss diets. Much of the data are diet correlates (DTA values), but diets are almost never mentioned, except in the Methods. For example, the authors say: "An overall shift towards increased dental topographic trait magnitudes ..." (L137). Does that mean there was a shift toward increased herbivory? If so, why not mention the dietary shift? And if most of the sampled taxa are herbivores (see above comment), then shouldn't herbivory be a focal point of the paper?
(b) The authors should expand on "we used dentitions as ecological indicators" (L75). For non-experts, how/why are dentitions linked to ecology? And, again, why not mention diet? A strong link between tooth shape and diet is a critical assumption here (and one I'm sure that all mammalogists agree with), but the authors don't provide justification (at least in the Introduction) for that assumption. Many relevant papers cited later in the Methods could be cited in the Introduction (e.g., Evans et al. 2007).
(c) Include a better introduction of the sample, such as explicitly stating that your sample only includes placentals (assuming that's the case) and is focused on three major clades. Are non-placentals like multituberculates or stem placentals/eutherians found at Chinese Paleocene fossil localities and not sampled in the study, or are they absent in the sampled area?
(d) The way in which "integration" is being used should be defined. That is a loaded term which has been defined in different ways. I also recommend providing more explanation on the integration analyses and what the results mean.
If the authors don't have space to expand the main text, then they should at least expand on the topics in the supplement, with appropriate citations to the supplement in the main text.
(4) Finally, I'm not convinced that the results fully support the 'brawn before bite' hypothesis. I like the hypothesis. However, the 'brawn before ...' part of the hypothesis assumes that body size disparity (L63) increased first, and I don't think that pattern is ever shown. First, body size disparity is never reported or plotted (at least that I could find) - the authors just show the violin plots of the body sizes (Figures 1B, S6A). Second, the authors don't show evidence of an actual increase in body size disparity. Instead, they seem to assume that there was a rapid diversification in the earliest Paleocene, and thus the early Paleocene bin has already "reached maximum saturation" (L148). But what if the body size disparity in the latest Cretaceous was the same as that in the Paleocene? (Although that's unlikely, note that papers like Clauset & Redner 2009 and Grossnickle & Newham 2016 found evidence of greater body size disparity in the latest Cretaceous than is commonly recognized.) Similarly, what if body size disparity increased rapidly in the Eocene? Wouldn't that suggest a 'BITE before brawn' hypothesis? So, without showing when an increase in body size diversity occurred, I don't think that the authors can make a strong argument for 'brawn before [insert any trait]".
Although it's probably well beyond the scope of the study to add Cretaceous or Eocene data, the authors could at least review literature on body size patterns during those times to provide greater evidence for an earliest Paleocene increase in size disparity.
-
Author response:
eLife Assessment
This important study fills a major geographic and temporal gap in understanding Paleocene mammal evolution in Asia and proposes an intriguing "brawn before bite" hypothesis grounded in diverse analytical approaches. However, the findings are incomplete because limitations in sampling design - such as the use of worn or damaged teeth, the pooling of different tooth positions, and the lack of independence among teeth from the same individuals - introduce uncertainties that weaken support for the reported disparity patterns. The taxonomic focus on predominantly herbivorous clades also narrows the ecological scope of the results. Clarifying methodological choices, expanding the ecological context, and tempering evolutionary interpretations would substantially strengthen the study.
We thank Dr. Rasmann for the constructive evaluation of our manuscript. Considering the reviewers’ comments, we plan to implement revisions to our study focusing on (1) expansion of the fossil sample description, including a detailed account of the process of excluding extremely worn or damaged teeth from all analyses, (2) expanded reporting of the analyses done on individual tooth positions, and tempering the interpretation of the pooled samples in light of the issues raised by reviewers, (3) providing a more comprehensive introduction that includes an overview of the Paleocene mammal faunas in south China, which unevenly samples certain clades whereas others are extremely rare, and why the current available fossil samples would not permit a whole-fauna analysis to be adequately conducted across the three land mammal age time bins of the Paleocene in China. We believe these revisions would substantially strengthen the study’s robustness and impact for understanding the ecomorphological evolution of the earliest abundant placental mammals during the Paleocene in Asia.
Public Reviews:
Reviewer #1 (Public review):
Summary:
This work provides valuable new insights into the Paleocene Asian mammal recovery and diversification dynamics during the first ten million years post-dinosaur extinction. Studies that have examined the mammalian recovery and diversification post-dinosaur extinction have primarily focused on the North American mammal fossil record, and it's unclear if patterns documented in North America are characteristic of global patterns. This study examines dietary metrics of Paleocene Asian mammals and found that there is a body size disparity increase before dietary niche expansion and that dietary metrics track climatic and paleobotanical trends of Asia during the first 10 million years after the dinosaur extinction.
Strengths:
The Asian Paleocene mammal fossil record is greatly understudied, and this work begins to fill important gaps. In particular, the use of interdisciplinary data (i.e., climatic and paleobotanical) is really interesting in conjunction with observed dietary metric trends.
Weaknesses:
While this work has the potential to be exciting and contribute greatly to our understanding of mammalian evolution during the first 10 million years post-dinosaur extinction, the major weakness is in the dental topographic analysis (DTA) dataset.
There are several specimens in Figure 1 that have broken cusps, deep wear facets, and general abrasion. Thus, any values generated from DTA are not accurate and cannot be used to support their claims. Furthermore, the authors analyze all tooth positions at once, which makes this study seem comprehensive (200 individual teeth), but it's unclear what sort of noise this introduces to the study. Typically, DTA studies will analyze a singular tooth position (e.g., Pampush et al. 2018 Biol. J. Linn. Soc.), allowing for more meaningful comparisons and an understanding of what value differences mean. Even so, the dataset consists of only 48 specimens. This means that even if all the specimens were pristinely preserved and generated DTA values could be trusted, it's still only 48 specimens (representing 4 different clades) to capture patterns across 10 million years. For example, the authors note that their results show an increase in OPCR and DNE values from the middle to the late Paleocene in pantodonts. However, if a singular tooth position is analyzed, such as the lower second molar, the middle and late Paleocene partitions are only represented by a singular specimen each. With a sample size this small, it's unlikely that the authors are capturing real trends, which makes the claims of this study highly questionable.
We thank Reviewer 1 for their careful review of our manuscript. A major external limitation of the application of DTA to fossil samples is the availability of specimens. Whereas a typical study design using extant or geologically younger/more abundant fossil species would preferably sample much larger quantities of teeth from each treatment group (time bins, in our case), the rarity of well-preserved Paleocene mammalian dentitions in Asia necessitates the analysis of small samples in order to make observations regarding major trends in a region and time period otherwise impossible to study (see Chow et al. 1977). That said, we plan to clarify methodological details in response to the reviewer’s comments, including a more comprehensive explanation of our criteria for exclusion of broken tooth crowns from the analyses. We also plan to expand our results reporting on individual tooth position analysis, potentially including resampling and/or simulation analyses to assess the effect of small and uneven samples on our interpretation of results. Lastly, we plan to revise the discussion and conclusion accordingly, including more explicit distinction between well-supported findings that emerge from various planned sensitivity analyses, versus those that are more speculative and tentative in nature.
Chow, M., Zhang, Y., Wang, B., and Ding, S. (1977). Paleocene mammalian fauna from the Nanxiong Basin, Guangdong Province. Paleontol. Sin. New Ser. C 20, 1–100.
Reviewer #2 (Public review):
Summary:
This study uses dental traits of a large sample of Chinese mammals to track evolutionary patterns through the Paleocene. It presents and argues for a 'brawn before bite' hypothesis - mammals increased in body size disparity before evolving more specialized or adapted dentitions. The study makes use of an impressive array of analyses, including dental topographic, finite element, and integration analyses, which help to provide a unique insight into mammalian evolutionary patterns.
Strengths:
This paper helps to fill in a major gap in our knowledge of Paleocene mammal patterns in Asia, which is especially important because of the diversification of placentals at that time. The total sample of teeth is impressive and required considerable effort for scanning and analyzing. And there is a wealth of results for DTA, FEA, and integration analyses. Further, some of the results are especially interesting, such as the novel 'brawn before bite' hypothesis and the possible link between shifts in dental traits and arid environments in the Late Paleocene. Overall, I enjoyed reading the paper, and I think the results will be of interest to a broad audience.
Weaknesses:
I have four major concerns with the study, especially related to the sampling of teeth and taxa, that I discuss in more detail below. Due to these issues, I believe that the study is incomplete in its support of the 'brawn before bite' hypothesis. Although my concerns are significant, many of them can be addressed with some simple updates/revisions to analyses or text, and I try to provide constructive advice throughout my review.
(1) If I understand correctly, teeth of different tooth positions (e.g., premolars and molars), and those from the same specimen, are lumped into the same analyses. And unless I missed it, no justification is given for these methodological choices (besides testing for differences in proportions of tooth positions per time bin; L902). I think this creates some major statistical concerns. For example, DTA values for premolars and molars aren't directly comparable (I don't think?) because they have different functions (e.g., greater grinding function for molars). My recommendation is to perform different disparity-through-time analyses for each tooth position, assuming the sample sizes are big enough per time bin. Or, if the authors maintain their current methods/results, they should provide justification in the main text for that choice.
We thank Reviewer 2 for raising several issues worthy of clarification. Separate analyses for individual tooth positions were performed but not emphasized in the first version of the study. In our revised version we plan to highlight the nuances of the results from premolar versus molar partition analyses.
Also, I think lumping teeth from the same specimen into your analyses creates a major statistical concern because the observations aren't independent. In other words, the teeth of the same individual should have relatively similar DTA values, which can greatly bias your results. This is essentially the same issue as phylogenetic non-independence, but taken to a much greater extreme.
It seems like it'd be much more appropriate to perform specimen-level analyses (e.g., Wilson 2013) or species-level analyses (e.g., Grossnickle & Newham 2016) and report those results in the main text. If the authors believe that their methods are justified, then they should explain this in the text.
We plan to emphasize individual tooth position analyses in our revisions, and provide a stronger justification for our current treatment of multiple teeth from the same individual specimens as independent samples. We recognize the statistical nonindependence raised by Reviewer 2, but we would point out that from an ecomorphological perspective, it is unclear to us that the heterodont dentition of these early Cenozoic placental mammals should represent a single ecological signal (and thus warrant using only a single tooth position as representative of an individual’s DTA values). We plan to closely examine the nature of nonindependence in the DTA data within individuals, to assess a balanced approach to maximize information content from the relatively small and rare fossil samples used, while minimizing signal nonindependence across the dentition.
(2) Maybe I misunderstood, but it sounds like the sampling is almost exclusively clades that are primarily herbivorous/omnivorous (Pantodonta, Arctostylopida, Anagalida, and maybe Tillodonta), which means that the full ecomorphological diversity of the time bins is not being sampled (e.g., insectivores aren't fully sampled). Similarly, the authors say that they "focused sampling" on those major clades and "Additional data were collected on other clades ... opportunistically" (L628). If they favored sampling of specific clades, then doesn't that also bias their results?
If the study is primarily focused on a few herbivorous clades, then the Introduction should be reframed to reflect this. You could explain that you're specifically tracking herbivore patterns after the K-Pg.
We plan to revise the introduction section to more accurately reflect the emphasis on those clades. However, we would note that conventional dietary ecomorphology categories used to characterize later branching placental mammals are likely to be less informative when applied to their Paleocene counterparts. Although there are dental morphological traits that began to characterize major placental clades during the Paleocene, distinctive dietary ecologies have not been demonstrated for most of the clade representatives studied. Thus, insectivory was probably not restricted to “Insectivora”, nor carnivory to early Carnivmorpha or “Creodonta”, each of which represented less than 5% of the taxonomic richness during the Paleocene in China (Wang et al. 2007).
Wang, Y., Meng, J., Ni, X., and Li, C. (2007). Major events of Paleogene mammal radiation in China. Geol. J. 42, 415–430.
(3) There are a lot of topics lacking background information, which makes the paper challenging to read for non-experts. Maybe the authors are hindered by a short word limit. But if they can expand their main text, then I strongly recommend the following:
(a) The authors should discuss diets. Much of the data are diet correlates (DTA values), but diets are almost never mentioned, except in the Methods. For example, the authors say: "An overall shift towards increased dental topographic trait magnitudes ..." (L137). Does that mean there was a shift toward increased herbivory? If so, why not mention the dietary shift? And if most of the sampled taxa are herbivores (see above comment), then shouldn't herbivory be a focal point of the paper?
We plan to revise the text to make clearer connections between DTA and dietary inferences, and at the same time advise caution in making one-to-one linkages between them. Broadly speaking, dental indices such as DTA are phenotypic traits, and as in other phenotypic traits, the strength of structure-function relationships needs to be explicitly established before dietary ecological inferences can be confidently made. There is, to date, no consistent connection between dental topology and tooth use proxies and biomechanical traits in extant non-herbivorous species (e.g., DeSantis et al. 2017, Tseng and DeSantis 2024), and in our analyses, FEA and DTA generally did not show strong correlations to each other. Thus, we plan to continue to exercise care in interpreting DTA data as dietary data.
DeSantis LRG, Tseng ZJ, Liu J, Hurst A, Schubert BW, Jiangzuo Q. Assessing niche conservatism using a multiproxy approach: dietary ecology of extinct and extant spotted hyenas. Paleobiology. 2017;43(2):286-303. doi:10.1017/pab.2016.45
Tseng ZJ, DeSantis LR. Relationship between tooth macrowear and jaw morphofunctional traits in representative hypercarnivores. PeerJ. 2024 Nov 11;12:e18435.
(b) The authors should expand on "we used dentitions as ecological indicators" (L75). For non-experts, how/why are dentitions linked to ecology? And, again, why not mention diet? A strong link between tooth shape and diet is a critical assumption here (and one I'm sure that all mammalogists agree with), but the authors don't provide justification (at least in the Introduction) for that assumption. Many relevant papers cited later in the Methods could be cited in the Introduction (e.g., Evans et al. 2007).
Thank you for this suggestion. We plan to expand the introduction section to better contextualize the methodological basis for the work presented.
(c) Include a better introduction of the sample, such as explicitly stating that your sample only includes placentals (assuming that's the case) and is focused on three major clades. Are non-placentals like multituberculates or stem placentals/eutherians found at Chinese Paleocene fossil localities and not sampled in the study, or are they absent in the sampled area?
We thank Reviewer 2 for raising this important point worthy of clarification. Multituberculates are completely absent from the first two land mammal ages in the Paleocene of Asia, and non-placentals are rare in general (Wang et al. 2007). We plan to provide more context for the taxonomic sampling choices made in the study.
Wang, Y., Meng, J., Ni, X., and Li, C. (2007). Major events of Paleogene mammal radiation in China. Geol. J. 42, 415–430.
(d) The way in which "integration" is being used should be defined. That is a loaded term which has been defined in different ways. I also recommend providing more explanation on the integration analyses and what the results mean.
If the authors don't have space to expand the main text, then they should at least expand on the topics in the supplement, with appropriate citations to the supplement in the main text.
We plan to clarify our usage of “integration” to enable readers to accurately interpret what we mean by it.
(4) Finally, I'm not convinced that the results fully support the 'brawn before bite' hypothesis. I like the hypothesis. However, the 'brawn before ...' part of the hypothesis assumes that body size disparity (L63) increased first, and I don't think that pattern is ever shown. First, body size disparity is never reported or plotted (at least that I could find) - the authors just show the violin plots of the body sizes (Figures 1B, S6A). Second, the authors don't show evidence of an actual increase in body size disparity. Instead, they seem to assume that there was a rapid diversification in the earliest Paleocene, and thus the early Paleocene bin has already "reached maximum saturation" (L148). But what if the body size disparity in the latest Cretaceous was the same as that in the Paleocene? (Although that's unlikely, note that papers like Clauset & Redner 2009 and Grossnickle & Newham 2016 found evidence of greater body size disparity in the latest Cretaceous than is commonly recognized.) Similarly, what if body size disparity increased rapidly in the Eocene? Wouldn't that suggest a 'BITE before brawn' hypothesis? So, without showing when an increase in body size diversity occurred, I don't think that the authors can make a strong argument for 'brawn before [insert any trait]".
Although it's probably well beyond the scope of the study to add Cretaceous or Eocene data, the authors could at least review literature on body size patterns during those times to provide greater evidence for an earliest Paleocene increase in size disparity.
We plan to provide a broader discussion and any supporting evidence from the Cretaceous and Eocene to either make a stronger case for “brawn before bite”, or to refine what we mean by brawn/size/size disparity.
-
-
www.densediscovery.com www.densediscovery.com
-
System design is the process of defining a system’s architecture, components, interfaces, and interactions in a structured way so the whole system meets its goals and requirements.
Put another way:
it’s planning how parts fit and work together, not just building them;
it creates a blueprint showing how elements communicate, behave, and support the system’s purpose;
it ensures the system will perform, scale, and remain reliable as conditions change.
System design is about setting constraints, incentives, and feedback so that any reasonable behavior leads to acceptable outcomes — without prescribing each individual action.
Design the conditions, not the conduct.
-
-
answers.childrenshospital.org answers.childrenshospital.org
-
Correlating with the EEGs, Dr. Law and her colleagues found that with every hour increase in average screen time, the children had more difficulties with attention and struggled more with executive functioning. However, because screen time is just one aspect of an infant’s environment, it is likely that multiple factors come into play, such as the quality of time with parents, the researchers say. It’s also possible that more active infants unintentionally receive more screen time as their parents try to manage their daily routines.
impotant evadance
-
-
yiamas.myflodesk.com yiamas.myflodesk.com
-
Because what if it's ok to be a bit woo-woo and also want to dive deeper into Aristotle? What if it's ok to be suspicious of all that IG manifestation talk, but still want to turn up the volume on the magic you know exists in the world? And finally, what if it's ok to be a bit nerdy, but also super curious about working with Greek and Balkan protection magic?
I think we asked a ton of hypothetical questions up top, so for sake of changing the structure up a bit I'd phrase this like:
This course assumes you're the kind of person who wants BOTH: the philosophical framework and the practical magic. Who finds TikTok manifestation culture too shallow but also knows there's something real happening when you work with ritual. Who wants to understand the 'why' behind the protection practices, not just follow instructions. If that's you, you're in the right place
-
Three things
just make this a little clearer -- "But maybe the problem isn't your self-help book addiction — it's the fact that you're trying to "fix" your social media hygiene at all. Consider this: 1... "
-
-
unpublishablepapers.substack.com unpublishablepapers.substack.com
-
**Summary ** 1) This isn’t nostalgia — it’s a structural change in childhood space
The essay argues that across history and cultures, kids have naturally carved out autonomous zones (streets, empty lots, forests, corners of towns) where they own time and space away from adults. That’s not a random pattern — it’s deeply human behavior. The Browser
The disappearance of these spaces isn’t just kids playing less. It’s a loss of a psychological environment where children make sense of the world on their own terms.
Insight: It reframes the problem from “kids spend more time inside” to “children are being structurally excluded from public life,” not by kids’ choices, but by how adult society is organized.
2) The cause is more built environment + social patterns than screens
The author pushes back against the common idea that the internet is the big culprit. Instead, he points to car-dependent suburbs, families spread far apart, and modern work patterns (parents not at home, schedules tightly managed), making free interaction physically harder. aman.bh
Insight: Technology is a symptom of isolation, not the root cause. The real bottlenecks are:
towns designed without gathering places
kids physically separated from peers
reliance on cars over walking/biking
3) Modern “play” is not truly play
There’s a distinction made between:
Structured activities (sports practice, classes with adults)
Unstructured peer play (kids deciding what to do, how to do it, together)
The latter is what’s disappearing. Organized activities fill time, but don’t create the same kind of autonomy and peer culture that spontaneous play does. aman.bh
Insight: If all your child’s social interactions are planned by adults, the dynamic changes — it becomes supervision, not co-participation.
4) Internet/online spaces are a child-managed arena
One reason kids gravitate online is because it’s one of the only unsupervised social spaces left. They aren’t free in the physical world, so they find agency where adults are less present (forums, chats, games). The Browser
New angle: The internet isn’t the cause of isolation — it’s a response to it. Kids go where they can control interactions without adult oversight.
5) The core issue isn’t “kids vs screens” — it’s where childhood autonomy can exist
This reframes the whole debate from blaming technologies to asking:
Where in the modern city can children act independently?
And the answer the essay hints at is: almost nowhere — so kids create their own spaces, even if imperfect.
Insight: Autonomy isn’t earned by limiting devices. It’s earned by restoring real-world environments where children can make choice, risk, negotiation, and friendship happen without adult orchestration.
6) Play functions as a designed culture, not an activity
When the essay references he “wishes children had forests,” he’s pointing to a deeper truth: What matters isn’t a physical object (forest) — it’s the freedom to explore, innovate, and improvise with peers.
Insight: Play loses value when it’s designed by adults for kids (e.g., programs, classes) and gains value when it’s designed by kids for themselves.
7) This problem isn’t just a “kids issue” — it’s a community design failure
The commentary makes it clear that the conditions limiting play — distance, traffic fears, suburban sprawl — are not random. They’re outcomes of how cities and societies organize:
roads instead of paths
fences instead of common spaces
schedules instead of unstructured time
Insight: If you want kids to have autonomy, you have to change the adult world — it’s not something kids can generate on their own.
-
-
papers.ssrn.com papers.ssrn.com
-
Author response:
The following is the authors’ response to the original reviews.
eLife Assessment
This Review Article explores the intricate relationship between humans and Mycobacterium tuberculosis (Mtb), providing an additional perspective on TB disease. Specifically, this review focuses on the utilization of systems-level approaches to study TB, while highlighting challenges in the frameworks used to identify the relevant immunologic signals that may explain the clinical spectrum of disease. The work could be further enhanced by better defining key terms that anchor the review, such as "unified mechanism" and "immunological route." This review will be of interest to immunologists as well as those interested in evolution and host-pathogen interactions.
We thank the editors for reviewing our article and for the primarily positive comments. We accept that better definition and terminology will improve the clarity of the message, and so have changed the wording as suggested above in the revised manuscript.
Public Reviews:
Reviewer #1 (Public review):
Summary:
This is an interesting and useful review highlighting the complex pathways through which pulmonary colonisation or infection with Mycobacterium tuberculosis (Mtb) may progress to develop symptomatic disease and transmit the pathogen. I found the section on immune correlates associated with individuals who have clearly been exposed to and reacted to Mtb but did not develop latent infections particularly valuable. However, several aspects would benefit from clarification.
Strengths:
The main strengths lie in the arguments presented for a multiplicity of immune pathways to TB disease.
Weaknesses:
The main weaknesses lie in clarity, particularly in the precise meanings of the three figures.
We accept this point, and have completely changed figure 2, and have expanded the legends for figure 1 and 3 to maximise clarity.
I accept that there is a 'goldilocks zone' that underpins the majority of TB cases we see and predominantly reflects different patterns of immune response, but the analogies used need to be more clearly thought through.
We are glad the reviewer agrees with the fundamental argument of different patterns of immunity, and have revised the manuscript throughout where we feel the analogies could be clarified.
Reviewer #2 (Public review):
Summary:
This is a thought-provoking perspective by Reichmann et al, outlining supportive evidence that Mycobacterium tuberculosis co-evolved with its host Homo Sapiens to both increase susceptibility to infection and reduce rates of fatal disease through decreased virulence. TB is an ancient disease where two modes of virulence are likely to have evolved through different stages of human evolution: one before the Neolithic Demographic Transition, where humans lived in sparse hunter-gatherer communities, which likely selected for prolonged Mtb infection with reduced virulence to allow for transmission across sparse populations. Conversely, following the agricultural and industrial revolutions, Mtb virulence is likely to have evolved to attack a higher number of susceptible individuals. These different disease modalities highlight the central idea that there are different immunological routes to TB disease, which converge on a disease phenotype characterized by high bacterial load and destruction of the extracellular matrix. The writing is very clear and provides a lot of supportive evidence from population studies and the recent clinical trials of novel TB vaccines, like M72 and H56. However, there are areas to support the thesis that have been described only in broad strokes, including the impact of host and Mtb genetic heterogeneity on this selection, and the alternative model that there are likely different TB diseases (as opposed to different routes to the same disease), as described by several groups advancing the concept of heterogeneous TB endotypes. I expand on specific points below.
Strengths:
The idea that Mtb evolved to both increase transmission (and possible commensalism with humans) with low rates of reactivation is intriguing. The heterogeneous TB phenotypes in the collaborative cross model (PMID: 35112666) support this idea, where some genetic backgrounds can tolerate a high bacterial load with minimal pathology, while others show signs of pathogenesis with low bacterial loads. This supports the idea that the underlying host state, driven by a number of factors like genetics and nutrition, is likely to explain whether someone will co-exist with Mtb without pathology, or progress to disease. I particularly enjoyed the discussion of the protective advantages provided by Mtb infection, which may have rewired the human immune system to provide protection against heterologous pathogens- this is supported by recent studies showing that Mtb infection provides moderate protection against SARS-CoV-2 (PMID: 35325013, and 37720210), and may have applied to other viruses that are likely to have played a more significant role in the past in the natural selection of Homo Sapiens.
We thank the reviewer for their positive comments, and also for pointing out work that we have overlooked citing previously. We now discuss and cite the work above as suggested
Modeling from Marcel Behr and colleagues (PMID: 31649096) indeed suggests that there are at least TB clinical phenotypes that likely mirror the two distinct phases of Mtb co-evolution with humans. Most of the TB disease progression occurs rapidly (within 1-2 years of exposure), and the rest are slow cases of reactivation over time. I enjoyed the discussion of the difference between the types of immune hits needed to progress to disease in the two scenarios, where you may need severe immune hits for rapid progression, a phenotype that likely evolved after the Neolithic transition to larger human populations. On the other hand, a series of milder immune events leading to reactivation after a long period of asymptomatic infection likely mirrors slow progression in the hunter-gatherer communities, to allow for prolonged transmission in scarce populations. Perhaps a clearer analysis of these models would be helpful for the reader.
We agree that we did not present these concepts in as much detail as we should, and so we now discuss this more on lines 81 – 83 and 184 - 187)
Weaknesses:
The discussion of genetic heterogeneity is limited and only discusses evidence from MSMD studies. Genetics is an important angle to consider in the co-evolution of Mtb and humans. There is a large body of literature on both host and Mtb genetic associations with TB disease. The very fact that host variants in one population do not necessarily cross-validate across populations is evidence in support of population-specific adaptations. Specific Mtb lineages are likely to have co-evolved with distinct human populations. A key reference is missing (PMID: 23995134), which shows that different lineages co-evolved with human migrations. Also, meta-analyses of human GWAS studies to define variants associated with TB are very relevant to the topic of co-evolution (e.g., PMID: 38224499). eQTL studies can also highlight genetic variants associated with regulating key immune genes involved in the response to TB. The authors do mention that Mtb itself is relatively clonal with ~2K SNPs marking Mtb variation, much of which has likely evolved under the selection pressure of modern antibiotics. However, some of this limited universe of variants can still explain co-adaptations between distinct Mtb lineages and different human populations, as shown recently in the co-evolution of lineage 2 with a variant common in Peruvians (PMID: 39613754).
We thank the reviewer for these comments and agree we failed to cite and discuss the work from Sebastian Gagneux’s group on co-migration, which we now discuss. We include a new paragraph discussing co-evolution as suggested on lines 145 – 155 and 218 -220 , citing the work proposed, which we agree enhances the arguments about co-evolution.
Although the examples of anti-TNF and anti-PD1 treatments are relevant as drivers of TB in limited clinical contexts, the bigger picture is that they highlight major distinct disease endotypes. These restricted examples show that TB can be driven by immune deficiency (as in the case of anti-TNF, HIV, and malnutrition) or hyperactivation (as in the case of anti-PD1 treatment), but there are still certainly many other routes leading to immune suppression or hyperactivation. Considering the idea of hyper-activation as a TB driver, the apparent higher rate of recurrence in the H56 trial referenced in the review is likely due to immune hyperactivation, especially in the context of residual bacteria in the lung. These different TB manifestations (immune suppression vs immune hyperactivation) mirror TB endotypes described by DiNardo et al (PMID: 35169026) from analysis of extensive transcriptomic data, which indicate that it's not merely different routes leading to the same final endpoint of clinical disease, but rather multiple different disease endpoints. A similar scenario is shown in the transcriptomic signatures underlying disease progression in BCG-vaccinated infants, where two distinct clusters mirrored the hyperactivation and immune suppression phenotypes (PMID: 27183822). A discussion of how to think about translating the extensive information from system biology into treatment stratification approaches, or adjunct host-directed therapies, would be helpful.
We agree with the points made and that the two publications above further enhance the paper. We have added discussion of the different disease endpoints on line 65 - 67, the evidence regarding immune herpeactivation versus suppression in the vaccination study on lines 162 - 164, and expanded on the translational implications on lines 349 – 352.
Reviewer #3 (Public review):
Summary:
This perspective article by Reichmann et al. highlights the importance of moving beyond the search for a single, unified immune mechanism to explain host-Mtb interactions. Drawing from studies in immune profiling, host and bacterial genetics, the authors emphasize inconsistencies in the literature and argue for broader, more integrative models. Overall, the article is thought-provoking and well-articulated, raising a concept that is worth further exploration in the TB field.
Strengths:
Timely and relevant in the context of the rapidly expanding multi-omics datasets that provide unprecedented insights into host-Mtb interactions.
Weaknesses (Minor):
Clarity on the notion of a "unified mechanism". It remains unclear whether prior studies explicitly proposed a single unifying immunological model. While inconsistencies in findings exist, they do not necessarily demonstrate that earlier work was uniformly "single-minded". Moreover, heterogeneity in TB has been recognized previously (PMIDs: 19855401, 28736436), which the authors could acknowledge.
We accept this point and have toned down the language, acknowledging that we are expanding on an argument that others have made, whilst focusing on the implications for the systems immunology era, and cite the previous work as suggested.
Evolutionary timeline and industrial-era framing. The evolutionary model is outdated. Ancient DNA studies place the Mtb's most recent common ancestor at ~6,000 years BP (PMIDs: 25141181; 25848958). The Industrial Revolution is cited as a driver of TB expansion, but this remains speculative without bacterial-genomics evidence and should be framed as a hypothesis. Additionally, the claim that Mtb genomes have been conserved only since the Industrial Revolution (lines 165-167) is inaccurate; conservation extends back to the MRCA (PMID: 31448322).
Our understanding is that the evolutionary timeline is not fully resolved, with conflicting evidence proposing different dates. The ancient DNA studies giving a timeline of 6,000 years seem to oppose the evidence of evidence of Mtb infection of humans in the middle east 10,000 years ago, and other estimates suggesting 70,000 years. Therefore, we have cited the work above and added a sentence highlighting that different studies propose different timelines. We would propose the industrial revolution created the ideal societal conditions for the expansion of TB, and this would seem widely accepted in the field, but have added a proviso as suggested. We did not intent to claim that Mtb genomes have been conserved since the industrial revolution, the point we were making is that despite rapid expansion within human populations, it has still remained conserved. We therefore have revised our discussion of the conservation of the Mtb genomes on lines and 72 – 74, 81 – 83 and 185 – 190.
Trained immunity and TB infection. The treatment of trained immunity is incomplete. While BCG vaccination is known to induce trained immunity (ref 59), revaccination does not provide sustained protection (ref 8), and importantly, Mtb infection itself can also impart trained immunity (PMID: 33125891). Including these nuances would strengthen the discussion.
We have refined this section. We did cite PMID: 33125891 in the original submission but have changed the wording to emphasise the point on line …
Recommendations for the authors:
Reviewer #1 (Recommendations for the authors):
Abstract
Line 30: What is an immunological route? Suggest
”...host-pathogen interaction, with diverse immunological processes leading to TB disease (10%) or stable lifelong association or elimination. We suggest these alternate relationships result from the prolonged co-evolution of the pathogen with humans and may even confer a survival advantage in the 90% of exposures that do not progress to disease.”
Thank you, we have reworded the abstract along the lines suggested above, but not identically to allow for other reviewer comments.
Introduction
Ln 43: It is misleading to suggest that the study of TB was the leading influence in establishing the Koch's postulates framework. Many other infections were involved, and Jacob Henle, one of Koch's teachers, is credited with the first clear formulation (see Evans AS. 1976 THE YALE JOURNAL OF BIOLOGY AND MEDICIN PMID: 782050).
We have downplayed the language, stating that TB “contributed” to the formulation if Koch’s postulated.
Ln 46: While the review rightly emphasises intracellular infection in macrophages, the importance and abundance of extracellular bacilli should not be ignored, particularly in transmission and in cavities.
We agree, and have added text on the importance of extracellular bacteria and transmission.
Ln: 56: This is misleading as primary disease prevention is implied, whereas the vaccine was given to individuals presumed to be already infected (TST or IGRA positive). Suggest ..."reduces by 50% progression to overt TB disease when given to those with immunological evidence of latent infection.
Thank you, edit made as suggested
Ln 62: Not sure why it is urgent. Suggest "high priority".
Wording changed as suggested.
Figure 1 needs clarification. The colour scale appears to signify the strength or vigour of the immune response so that disease is associated with high (orange/red) or low (green/blue) activity. The arrows seem to imply either a sequence or a route map when all we really have is an association with a plausible mechanistic link. They might also be taken to imply a hierarchy that is not appropriate. I'm not sure that the X-rays and arrows add anything, and the rectangle provides the key information on its own. Clarify please.
We have clarified the figure legend. We feel the X-rays give the clinical context, and so have kept them, and now state in the legend that this is highlighting that there are diverse pathways leading to active disease to try to emphasise the point the figure is illustrating.
Ln 149-157: I agree that the current dogma is that overt pulmonary disease is required to spread Mtb and fuel disease prevalence. It is vitally important to distinguish the spread of the organism from the occurrence of disease (which does not, of itself, spread). However, both epidemiological (e.g. Ryckman TS, et al. 2022Proc Natl Acad Sci U S A:10.1073/pnas.2211045119) and recent mechanistic (Dinkele R, et al. 2024iScience:10.1016/j.isci.2024.110731, Patterson B, et al. 2024Proc Natl Acad Sci U S A:10. E1073/pnas.2314813121, Warner DF, et al. 2025Nat Rev Microbiol:10.1038/s41579-025-01201-x) studies indicate the importance of asymptomatic infections, and those associated with sputum positivity have recently been recognised by WHO. I think it will be important to acknowledge the importance of this aspect and consider how immune responses may or may not contribute. I regard the view that Mtb is an obligate pathogen, dependent on overt pTB for transmission, as needing to be reviewed.
We agree that we did not give sufficient emphasis to the emerging evidence on asymptomatic infections, and that this may play an important part in transmission in high incidence settings. We now include a discussion on this, and citation of the papers above, on lines 168 – 170.
Ln 159: The terms colonise and colonisation are used, without a clear definition, several times. My view is that both refer to the establishment and replication of an organism on or within a host without associated damage. Where there is associated damage, this is often mediated by immune responses. In this header, I think "establishment in humanity" would be appropriate.
We agree with this point and have changed the header as suggested, and clarified our meaning when we use the term colonisation, which the reviewer correctly interprets.
Ln 181-: I strongly support the view that Mtb has contributed to human selection, even to the suggestion that humanity is adapted to maintain a long-term relationship with Mtb
Thank you, and we have expanded on this evidence as suggested by other reviewers.
Ln 189: improved.
Apologies, typo corrected.
Figure 2: I was also confused by this. The x-axis does not make sense, as a single property should increase. Moreover, does incidence refer to incidence in individuals with that specific balance of resistance and susceptibility, or contribution to overall global incidence - I suspect the latter (also, prevalence would make more sense). At the same time, the legend implies that those with high resistance to colonisation will be infrequent in the population, suggesting that the Y axis should be labelled "frequency in human population". Finally, I can't see what single label could apply to the X axis. While the implication that the majority of global infections reflect a balance between the resistance and susceptibilities is indicated, a frequency distribution does not seem an appropriate representation.
The reviewer is correct that the X axis is aiming to represent two variables, which is not logical, and so we have completely changed this figure to a simple one that we hope makes the point clearly and have amended the legend appropriately. We are aiming to highlight the selective pressures of Mtb on the human population over millennia.
Ln 244: Immunological failure - I agree with the statement but again find the figure (3) unhelpful. Do we start or end in the middle? Is the disease the outside - if so, why are different locations implied? The notion of a maze has some value, but the bacteria should start and finish in the same place by different routes.
We are attempting to illustrate the concept that escape from host immunological control can occur through different mechanisms. As this comment was just from one reviewer, we have left the figure unchanged but have expanded the legend to try to make the point that this is just a conceptual illustration of multiple routes to disease.
Ln 262 onward: I broadly agree with the points made about omic technologies, but would wish to see major emphasis on clear phenotyping of cases. There is something of a contradiction in the review between the emphasis on the multiplicity of immunological processes leading ultimately to disease and the recommendation to analyse via omics, which, in their most widely applied format, bundle these complexities into analyses of the humoral and cellular samples available in blood. Admittedly, the authors point out opportunities for 3-dimensional and single-cell analyses, but it is difficult to see where these end without extrapolation ad infinitum.
We totally agree that clear phenotyping of infection is critical, and expand on this further on lines 307 - 309.
Reviewer #2 (Recommendations for the authors):
I suggest expanding on the genetic determinants of Mtb/host co-evolution.
Thank you, we have now expanded on these sections as suggested.
Reviewer #3 (Recommendations for the authors):
We are in an era of exploding large-scale datasets from multi-omics profiling of Mtb and host interactions, offering an unprecedented lens to understand the complexity of the host immune response to Mtb-a pathogen that has infected human populations for thousands of years. The guiding philosophy for how to interpret this tremendous volume of data and what models can be built from it will be critical. In this context, the perspective article by Reichmann et al. raises an interesting concept: to "avoid unified immune mechanisms" when attempting to understand the immunology underpinning host-Mtb interactions. To support their arguments, the authors review studies and provide evidence from immune profiling, host and bacterial genetics, and showcase several inconsistencies. Overall, this perspective article is well articulated, and the concept is worthwhile for further exploration. A few comments for consideration:
Clarity on the notion of a "unified mechanism". Was there ever a single, clearly proposed unified immunological mechanism? For example, in lines 64-65, the authors criticize that almost all investigations into immune responses to Mtb are based on the premise that a unifying disease mechanism exists. However, after reading the article, it was not clear to me how previous studies attempted to unify the model or what that unifying mechanism was. While inconsistencies in findings certainly exist, they do not necessarily indicate that prior work was guided by a unified framework. I agree that interpreting and exploring data from a broader perspective is valuable, but I am not fully convinced that previous studies were uniformly "single-minded". In fact, the concept of heterogeneity in TB has been previously discussed (e.g., PMIDs: 19855401, 28736436).
We accept this point, and that we have overstated the argument and not acknowledged previous work sufficiently. We now downplay the language and cite the work as proposed.
However, we would propose that essentially all published studies imply that single mechanisms underly development of disease. The authors are not aware of any manuscript that concludes “Therefore, xxxx pathway is one of several that can lead to TB disease”, instead they state “Therefore, xxxx pathway leads to TB disease”. The implication of this language is that the mechanism described occurs in all patients, whilst in fact it likely only is involved in a subset. We have toned down the language and expand on this concept on line 268 – 270.
Evolutionary timeline and industrial-era framing. The evolutionary model needs updating. The manuscript cites a "70,000-year" origin for Mtb, but ancient-DNA studies place the most recent common ancestor at ~6,000 years BP (PMIDs: 25141181; 25848958). The Industrial Revolution is invoked multiple times as a driver of TB expansion, yet the magnitude of its contribution remains debated and, to my knowledge, lacks direct bacterial-genomics evidence for causal attribution; this should be framed as a hypothesis rather than a conclusion. In addition, the statement in lines 165-167 is inaccurate: at the genome level, Mtb has remained highly conserved since its most recent common ancestor-not specifically since the Industrial Revolution (PMID: 31448322).
We accept these points and have made the suggested amendments, as outlined in the public responses. Our understanding is that the evidence about the most common ancestor is controversial; if the divergence of human populations occurred concurrently with Mtb, then this must have been significantly earlier than 6,000 years ago, and so there are conflicting arguments in this domain.
Trained immunity and TB infection. The discussion of trained immunity could be expanded. Reference 59 suggests the induction of innate immune training, but reference 8 reports that revaccination does not confer protection against sustained TB infection, indicating that at least "re"-vaccination may not enhance protection. Furthermore, while BCG is often highlighted as a prototypical inducer of trained immunity, real-world infection occurs through Mtb itself. Importantly, a later study demonstrated that Mtb infection can also impart trained immunity (PMID: 33125891). Integrating these findings would provide a more nuanced view of how both vaccination and infection shape innate immune training in the TB context.
We thank the reviewer for these suggestions and have edited the relevant section to include these studies.
Tags
Annotators
URL
-
-
www.biorxiv.org www.biorxiv.org
-
Reviewer #2 (Public review):
Summary:
This manuscript investigates how olfactory representations are transformed along the cortico-hippocampal pathway in mice during a non-associative learning paradigm involving novel and familiar odors. By recording single-unit activity in several key brain regions (AON, aPCx, LEC, CA1, and SUB), the authors aim to elucidate how stimulus identity and experience are encoded and how these representations change across the pathway.
The study addresses an important question in sensory neuroscience regarding the interplay between sensory processing and signaling novelty/familiarity. It provides insights into how the brain processes and retains sensory experiences, suggesting that the earlier stations in the olfactory pathway, the AON aPCx, play a central role in detecting novelty and encoding odor, while areas deeper into the pathway (LEC, CA1 & Sub) are more sparse and encodes odor identity but not novelty/familiarity. However, there are several concerns related to methodology, data interpretation, and the strength of the conclusions drawn.
Strengths:
The authors combine the use of modern tools to obtain high-density recordings from large populations of neurons at different stages of the olfactory system (although mostly one region at a time) with elegant data analyses to study an important and interesting question.
Weaknesses:
The first and biggest problem I have with this paper is that it is very confusing, and the results seem to be all over the place. In some parts, it seems like the AON and aPCx are more sensitive to novelty; in others, it seems the other way around. I find their metrics confusing and unconvincing. For example, the example cells in Figure 1C shows an AON neuron with a very low spontaneous firing rate and a CA1 with a much higher firing rate, but the opposite is true in Fig. 2A. So, what are we to make of Fig. 2C that shows the difference in firing rates between novel vs. familiar odors measured as a difference in spikes/sec. The meaning of this is unclear. The authors could have used a difference in Z-scored responses to normalize different baseline activity levels. (This is just one example of a problem with the methodology.)
There are a lot of high-level data analyses (e.g., decoding, analyzing decoding errors, calculating mutual information, calculating distances in state space, etc.) but very little neural data (except for Fig. 2C, and see my comment above about how this is flawed). So, if responses to novel vs. familiar odors are different in the AON and aPCx, how are they different? Why is decoding accuracy better for novel odors in CA1 but better for familiar odors in SUB (Fig. 3A)? The authors identify a small subset of neurons that have unusually high weights in the SVM analyses that contribute to decoding novelty, but they don't tell us which neurons these are and how they are responding differently to novel vs. familiar odors.
The authors call AON and aPCx "primary sensory cortices" and LEC, CA1, and Sub "multisensory areas". This is a straw man argument. For example, we now know that PCx encodes multimodal signals (Poo et al. 2021, Federman et al., 2024; Kehl et al., 2024), and LEC receives direct OB inputs, which has traditionally been the criterion for being considered a "primary olfactory cortical area". So, this terminology is outdated and wrong, and although it suits the authors' needs here in drawing distinctions, it is simplistic and not helpful moving forward.
Why not simply report z-scored firing rates for all neurons as a function of trial number? (e.g., Jacobson & Friedrich, 2018). Fig. 2C is not sufficient. For example, in the Discussion, they say, "novel stimuli caused larger increases in firing rates than familiar stimuli" (L. 270), but what does this mean? Odors typically increase the firing in some neurons and suppress firing in others. Where does the delta come from? Is this because novel odors more strongly activate neurons that increase their firing or because familiar odors more strongly suppress neurons?
Ls. 122-124 - If cells in AON and aPCx responded the same way to novel and familiar odors, then we would say that they only encode for odor and not at all for experience. So, I don't understand why the authors say these areas code for a "mixed representation of chemical identity and experience." "On the other hand," if LEC, CA1, and SUB are odor selective and only encode novel odors, then these areas, not AON and aPCx, are the jointly encoding chemical identity and experience. Also, I do not understand why, here, they say that AON and PCx respond to both while LEC, CA1, and SUB were selective for novel stimuli, but the authors then go on to argue that novelty is encoded in the AON and PCx, but not in the LEC, CA1, and SUB.
Ls. 132-140 - As presented in the text and the figure, this section is unclear and confusing. Their use of the word "shuffled" is a major source of this confusion, because this typically is the control that produces outcomes at chance level. More importantly, it seems as though they did the wrong analysis here. A better way to do this analysis is to train on some of the odors and test on an untrained odor (i.e., what Bernardi et al., 2021 called "cross-condition generalization performance"; CCGP).
Comments on revisions:
I think the authors have done an adequate job addressing the reviewers' concerns. Most importantly, I found the first version of the manuscript quite confusing, and the consequent clarifications have addressed this issue.
In several cases, I see their point, while I still disagree with whether they made the best decisions. However, the issues here do not fundamentally change the big-picture outcome, and if they want to dig in with their approaches (e.g., only using auROC or just reporting delta firing rates without any normalization), it's their choice.
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the current reviews.
Public Reviews:
Reviewer #1 (Public review):
Summary:
This study examines whether changes in pupil size index prediction-error-related updating during associative learning, formalised as information gain via Kullback-Leibler (KL) divergence. Across two independent tasks, pupil responses scaled with KL divergence shortly after feedback, with the timing and direction of the response varying by task. Overall, the work supports the view that pupil size reflects information-theoretic processes in a context-dependent manner.
Strengths:
This study provides a novel and convincing contribution by linking pupil dilation to informationtheoretic measures, such as KL divergence, supporting Zénon's hypothesis that pupil responses reflect information gain during learning. The robust methodology, including two independent datasets with distinct task structures, enhances the reliability and generalisability of the findings. By carefully analysing early and late time windows, the authors capture the timing and direction of prediction-error-related responses, oPering new insights into the temporal dynamics of model updating. The use of an ideal-learner framework to quantify prediction errors, surprise, and uncertainty provides a principled account of the computational processes underlying pupil responses. The work also highlights the critical role of task context in shaping the direction and magnitude of these ePects, revealing the adaptability of predictive processing mechanisms. Importantly, the conclusions are supported by rigorous control analyses and preprocessing sanity checks, as well as convergent results from frequentist and Bayesian linear mixed-ePects modelling approaches.
Weaknesses:
Some aspects of directionality remain context-dependent, and on current evidence cannot be attributed specifically to whether average uncertainty increases or decreases across trials. DiPerences between the two tasks (e.g., sensory modality and learning regime) limit direct comparisons of ePect direction and make mechanistic attribution cautious. In addition, subjective factors such as confidence were not measured and could influence both predictionerror signals and pupil responses. Importantly, the authors explicitly acknowledge these limitations, and the manuscript clearly frames them as areas for future work rather than settled conclusions.
Reviewer #2 (Public review):
Summary:
The authors investigate whether pupil dilation reflects information gain during associative learning, formalised as Kullback-Leibler divergence within an ideal observer framework. They examine pupil responses in a late time window after feedback and compare these to informationtheoretic estimates (information gain, surprise, and entropy) derived from two diPerent tasks with contrasting uncertainty dynamics.
Strength:
The exploration of task evoked pupil dynamics beyond the immediate response/feedback period and then associating them with model estimates was interesting and inspiring. This oPered a new perspective on the relationship between pupil dilation and information processing.
Weakness:
However, the interpretability of the findings remains constrained by the fundamental diPerences between the two tasks (stimulus modality, feedback type, and learning structure), which confound the claimed context-dependent ePects. The later time-window pupil ePects, although intriguing, are small in magnitude and may reflect residual noise or task-specific arousal fluctuations rather than distinct information-processing signals. Thus, while the study oPers valuable methodological insight and contributes to ongoing debates about the role of the pupil in cognitive inference, its conclusions about the functional significance of late pupil responses should be treated with caution.
Reviewer #3 (Public review):
Summary:
Thank you for inviting me to review this manuscript entitled "Pupil dilation oPers a time-window on prediction error" by Colizoli and colleagues. The study examines prediction errors, information gain (Kullback-Leibler [KL] divergence), and uncertainty (entropy) from an information-theory perspective using two experimental tasks and pupillometry. The authors aim to test a theoretical proposal by Zénon (2019) that the pupil response reflects information gain (KL divergence). The conclusion of this work is that (post-feedback) pupil dilation in response to information gain is context dependent.
Strengths:
Use of an established Bayesian model to compute KL divergence and entropy.
Pupillometry data preprocessing and multiple robustness checks.
Weaknesses:
Operationalization of prediction errors based on frequency, accuracy, and their interaction:
The authors rely on a more model-agnostic definition of the prediction error in terms of stimulus frequency ("unsigned prediction error"), accuracy, and their interaction ("signed prediction error"). While I see the point, I would argue that this approach provides a simple approximation of the prediction error, but that a model-based approach would be more appropriate.
Model validation:
My impression is that the ideal learner model should work well in this case. However, the authors don't directly compare model behavior to participant behavior ("posterior predictive checks") to validate the model. Therefore, it is currently unclear if the model-derived terms like KL divergence and entropy provide reasonable estimates for the participant data.
Lack of a clear conclusion:
The authors conclude that this study shows for the first time that (post-feedback) pupil dilation in response to information gain is context dependent. However, the study does not oPer a unifying explanation for such context dependence. The discussion is quite detailed with respect to taskspecific ePects, but fails to provide an overarching perspective on the context-dependent nature of pupil signatures of information gain. This seems to be partly due to the strong diPerences between the experimental tasks.
Recommendations for the authors:
Reviewer #1 (Recommendations for the authors):
I highly appreciate the care and detail in the authors' response and thank them for the ePort invested in revising the manuscript. They addressed the core concerns to a high standard, and the manuscript has substantially improved in methodological rigour (through additional controls/sanity checks and complementary mixed-ePects analyses) and in clarity of interpretation (by explicitly acknowledging context-dependence and tempering stronger claims). The present version reads clearly and is much strengthened overall. I only have a few minor points below:
Minor suggestions:
Abstract:
In the abstract KL is introduced as abbreviation, but at first occurence it should be written out as "Kullback-Leibler (KL)" for readers not familiar with it.
We thank the reviewer for catching this error. It has been correct in the version of record.
Methods:
I appreciate the additional bayesian LME analysis. I only had a few things that I thought were missing from knowing the parameters: 1) what was the target acceptance rate (default of .95?), 2) which family was used to model the response distribution: (default) "gaussian" or robust "student-t"? Depending on the data a student-t would be preferred, but since the author's checked the fit & the results corroborate the correlation analysis, using the default would also be fine! Just add the information for completeness.
Thank you for bringing this to our attention. We have now noted that default parameters were used in all cases unless otherwise mentioned.
Thank you once again for your time and consideration.
Reviewer #2 (Recommendations for the authors):
Thanks to the authors' ePort on revision. I am happy with this new version of manuscript.
Thank you once again for your time and consideration.
Reviewer #3 (Recommendations for the authors):
(1) Regarding comments #3 and #6 (first round) on model validation and posterior predictive checks, the authors replied that since their model is not a "generative" one, they can't perform posterior predictive checks. Crucially, in eq. 2, the authors present the p{tilde}^j_k variable denoting the learned probability of event k on trial j. I don't see why this can't be exploited for simulations. In my opinion, one could (and should) generate predictions based on this variable. The simplest implementation would translate the probability into a categorical choice (w/o fitting any free parameter). Based on this, they could assess whether the model and data are comparable.
We thank the reviewer for this clarification. The reviewer suggests using the probability distributions at each trial to predict which event should be chosen on each trial. More specifically, the event(s) with the highest probability on trial j could be used to generate a prediction for the choice of the participant on trial j. We agree that this would indeed be an interesting analysis. However, the response options of each task are limited to two-alternatives. In the cue-target task, four events are modeled (representing all possible cue-target conditions) while the participants’ response options are only “left” and “right”. Similarly, in the letter-color task, 36 events are modeled while the participants’ response options are “match” and “no-match”. In other words, we do not know which event (either four or 36, for the two tasks) the participant would have indicated on each trial. As an approximation to this fine-grained analysis, we investigated the relationship between the information-theoretic variables separately for error and correct trials. Our rationale was that we would have more insight into how the model fits depended on the participants’ actual behavior as compared with the ideal learner model.
(2) I recommend providing a plot of the linear mixed model analysis of the pupil data. Currently, results are only presented in the text and tables, but a figure would be much more useful.
We thank the reviewer for the suggestion to add a plot of the linear mixed model results. We appreciate the value of visualizing model estimates; however, we feel that the current presentation in the text and tables clearly conveys the relevant findings. For this reason, and to avoid further lengthening the manuscript, we prefer to retain the current format.
(3) I would consider only presenting the linear mixed ePects for the pupil data in the main results, and the correlation results in the supplement. It is currently quite long.
We thank the reviewer for this recommendation. We agree that the results section is detailed; however, we consider the correlation analyses to be integral to the interpretation of the pupil data and therefore prefer to keep them in the main text rather than move them to the supplement.
The following is the authors’ response to the original reviews
eLife Assessment
This important study seeks to examine the relationship between pupil size and information gain, showing opposite effects dependent upon whether the average uncertainty increases or decreases across trials. Given the broad implications for learning and perception, the findings will be of broad interest to researchers in cognitive neuroscience, decision-making, and computational modelling. Nevertheless, the evidence in support of the particular conclusion is at present incomplete - the conclusions would be strengthened if the authors could both clarify the differences between model-updating and prediction error in their account and clarify the patterns in the data.
Public Reviews:
Reviewer #1 (Public review):
Summary:
This study investigates whether pupil dilation reflects prediction error signals during associative learning, defined formally by Kullback-Leibler (KL) divergence, an information-theoretic measure of information gain. Two independent tasks with different entropy dynamics (decreasing and increasing uncertainty) were analyzed: the cue-target 2AFC task and the lettercolor 2AFC task. Results revealed that pupil responses scaled with KL divergence shortly after feedback onset, but the direction of this relationship depended on whether uncertainty (entropy) increased or decreased across trials. Furthermore, signed prediction errors (interaction between frequency and accuracy) emerged at different time windows across tasks, suggesting taskspecific temporal components of model updating. Overall, the findings highlight that pupil dilation reflects information-theoretic processes in a complex, context-dependent manner.
Strengths:
This study provides a novel and convincing contribution by linking pupil dilation to informationtheoretic measures, such as KL divergence, supporting Zénon's hypothesis that pupil responses reflect information gained during learning. The robust methodology, including two independent datasets with distinct entropy dynamics, enhances the reliability and generalisability of the findings. By carefully analysing early and late time windows, the authors capture the temporal dynamics of prediction error signals, offering new insights into the timing of model updates. The use of an ideal learner model to quantify prediction errors, surprise, and entropy provides a principled framework for understanding the computational processes underlying pupil responses. Furthermore, the study highlights the critical role of task context - specifically increasing versus decreasing entropy - in shaping the directionality and magnitude of these effects, revealing the adaptability of predictive processing mechanisms.
Weaknesses:
While this study offers important insights, several limitations remain. The two tasks differ significantly in design (e.g., sensory modality and learning type), complicating direct comparisons and limiting the interpretation of differences in pupil dynamics. Importantly, the apparent context-dependent reversal between pupil constriction and dilation in response to feedback raises concerns about how these opposing effects might confound the observed correlations with KL divergence.
We agree with the reviewer’s concerns and acknowledge that the speculation concerning the directional effect of entropy across trials can not be fully substantiated by the current study. As the reviewer points out, the directional relationship between pupil dilation and information gain must be due to other factors, for instance, the sensory modality, learning type, or the reversal between pupil constriction and dilation across the two tasks. Also, we would like to note that ongoing experiments in our lab already contradict our original speculation. In line with the reviewer’s point, we noted these differences in the section on “Limitations and future research” in the Discussion. To better align the manuscript with the above mentioned points, we have made several changes in the Abstract, Introduction and Discussion summarized below:
We have removed the following text from the Abstract and Introduction: “…, specifically related to increasing or decreasing average uncertainty (entropy) across trials.”
We have edited the following text in the Introduction (changes in italics) (p. 5):
“We analyzed two independent datasets featuring distinct associative learning paradigms, one characterized by increasing entropy and the other by decreasing entropy as the tasks progressed. By examining these different tasks, we aimed to identify commonalities (if any) in the results across varying contexts. Additionally, the contrasting directions of entropy in the two tasks enabled us to disentangle the correlation between stimulus-pair frequency and information gain in the postfeedback pupil response.
We have removed the following text from the Discussion:
“…and information gain in fact seems to be driven by increased uncertainty.”
“We speculate that this difference in the direction of scaling between information gain and the pupil response may depend on whether entropy was increasing or decreasing across trials.”
“…which could explain the opposite direction of the relationship between pupil dilation and information gain”
“… and seems to relate to the direction of the entropy as learning progresses (i.e., either increasing or decreasing average uncertainty).”
We have edited the following texts in the Discussion (changes in italics):
“For the first time, we show that the direction of the relationship between postfeedback pupil dilation and information gain (defined as KL divergence) was context dependent.” (p. 29):
Finally, we have added the following correction to the Discussion (p. 30):
“Although it is tempting to speculate that the direction of the relationship between pupil dilation and information gain may be due to either increasing or decreasing entropy as the task progressed, we must refrain from this conclusion. We note that the two tasks differ substantially in terms of design with other confounding variables and therefore cannot be directly compared to one another. We expand on these limitations in the section below (see Limitations and future research).”
Finally, subjective factors such as participants' confidence and internal belief states were not measured, despite their potential influence on prediction errors and pupil responses.
Thank you for the thoughtful comment. We agree with the reviewer that subjective factors, such as participants' confidence, can be important in understanding prediction errors and pupil responses. As per the reviewer’s point, we have included the following limitation in the Discussion (p. 33):
“Finally, while we acknowledge the potential relevance of subjective factors, such as the participants’ overt confidence reports, in understanding prediction errors and pupil responses, the current study focused on the more objective, model-driven measure of information-theoretic variables. This approach aligns with our use of the ideal learner model, which estimates information-theoretic variables while being agnostic about the observer's subjective experience itself. Future research is needed to explore the relationship between information-gain signals in pupil dilation and the observer’s reported experience of or awareness about confidence in their decisions.”
Reviewer #2 (Public review):
Summary:
The authors proposed that variability in post-feedback pupillary responses during the associative learning tasks can be explained by information gain, which is measured as KL divergence. They analysed pupil responses in a later time window (2.5s-3s after feedback onset) and correlated them with information-theory-based estimates from an ideal learner model (i.e., information gain-KL divergence, surprise-subjective probability, and entropy-average uncertainty) in two different associative decision-making tasks.
Strength:
The exploration of task-evoked pupil dynamics beyond the immediate response/feedback period and then associating them with model estimates was interesting and inspiring. This offered a new perspective on the relationship between pupil dilation and information processing.
Weakness:
However, disentangling these later effects from noise needs caution. Noise in pupillometry can arise from variations in stimuli and task engagement, as well as artefacts from earlier pupil dynamics. The increasing variance in the time series of pupillary responses (e.g., as shown in Figure 2D) highlights this concern.
It's also unclear what this complicated association between information gain and pupil dynamics actually means. The complexity of the two different tasks reported made the interpretation more difficult in the present manuscript.
We share the reviewer’s concerns. To make this point come across more clearly, we have added the following text to the Introduction (p. 5):
“The current study was motivated by Zenon’s hypothesis concerning the relationship between pupil dilation and information gain, particularly in light of the varying sources of signal and noise introduced by task context and pupil dynamics. By demonstrating how task context can influence which signals are reflected in pupil dilation, and highlighting the importance of considering their temporal dynamics, we aim to promote a more nuanced and model-driven approach to cognitive research using pupillometry.”
Reviewer #3 (Public review):
Summary:
This study examines prediction errors, information gain (Kullback-Leibler [KL] divergence), and uncertainty (entropy) from an information-theory perspective using two experimental tasks and pupillometry. The authors aim to test a theoretical proposal by Zénon (2019) that the pupil response reflects information gain (KL divergence). In particular, the study defines the prediction error in terms of KL divergence and speculates that changes in pupil size associated with KL divergence depend on entropy. Moreover, the authors examine the temporal characteristics of pupil correlates of prediction errors, which differed considerably across previous studies that employed different experimental paradigms. In my opinion, the study does not achieve these aims due to several methodological and theoretical issues.
Strengths:
(1) Use of an established Bayesian model to compute KL divergence and entropy.
(2) Pupillometry data preprocessing, including deconvolution.
Weaknesses:
(1) Definition of the prediction error in terms of KL divergence:
I'm concerned about the authors' theoretical assumption that the prediction error is defined in terms of KL divergence. The authors primarily refer to a review article by Zénon (2019): "Eye pupil signals information gain". It is my understanding that Zénon argues that KL divergence quantifies the update of a belief, not the prediction error: "In short, updates of the brain's internal model, quantified formally as the Kullback-Leibler (KL) divergence between prior and posterior beliefs, would be the common denominator to all these instances of pupillary dilation to cognition." (Zénon, 2019).
From my perspective, the update differs from the prediction error. Prediction error refers to the difference between outcome and expectation, while update refers to the difference between the prior and the posterior. The prediction error can drive the update, but the update is typically smaller, for example, because the prediction error is weighted by the learning rate to compute the update. My interpretation of Zénon (2019) is that they explicitly argue that KL divergence defines the update in terms of the described difference between prior and posterior, not the prediction error.
The authors also cite a few other papers, including Friston (2010), where I also could not find a definition of the prediction error in terms of KL divergence. For example [KL divergence:] "A non-commutative measure of the non-negative difference between two probability distributions." Similarly, Friston (2010) states: Bayesian Surprise - "A measure of salience based on the Kullback-Leibler divergence between the recognition density (which encodes posterior beliefs) and the prior density. It measures the information that can be recognized in the data." Finally, also in O'Reilly (2013), KL divergence is used to define the update of the internal model, not the prediction error.
The authors seem to mix up this common definition of the model update in terms of KL divergence and their definition of prediction error along the same lines. For example, on page 4: "KL divergence is a measure of the difference between two probability distributions. In the context of predictive processing, KL divergence can be used to quantify the mismatch between the probability distributions corresponding to the brain's expectations about incoming sensory input and the actual sensory input received, in other words, the prediction error (Friston, 2010; Spratling, 2017)."
Similarly (page 23): "In the current study, we investigated whether the pupil's response to decision outcome (i.e., feedback) in the context of associative learning reflects a prediction error as defined by KL divergence."
This is problematic because the results might actually have limited implications for the authors' main perspective (i.e., that the pupil encodes prediction errors) and could be better interpreted in terms of model updating. In my opinion, there are two potential ways to deal with this issue:
(a) Cite work that unambiguously supports the perspective that it is reasonable to define the prediction error in terms of KL divergence and that this has a link to pupillometry. In this case, it would be necessary to clearly explain the definition of the prediction error in terms of KL divergence and dissociate it from the definition in terms of model updating.
(b) If there is no prior work supporting the authors' current perspective on the prediction error, it might be necessary to revise the entire paper substantially and focus on the definition in terms of model updating.
We thank the reviewer for pointy out these inconsistencies in the manuscript and appreciate their suggestions for improvement. We take approach (a) recommended by the reviewer, and provide our reasoning as to why prediction error signals in pupil dilation are expected to correlate with information gain (defined as the KL divergence between posterior and prior belief distributions). This can be found in a new section in the introduction, copied here for convenience (p. 3-4):
“We reasoned that the link between prediction error signals and information gain in pupil dilation is through precision-weighting. Precision refers to the amount of uncertainty (inverse variance) of both the prior belief and sensory input in the prediction error signals [6,64–67]. More precise prediction errors receive more weighting, and therefore, have greater influence on model updating processes. The precisionweighting of prediction error signals may provide a mechanism for distinguishing between known and unknown sources of uncertainty, related to the inherent stochastic nature of a signal versus insufficient information of the part of the observer, respectively [65,67,68]. In Bayesian frameworks, information gain is fundamentally linked to prediction error, modulated by precision [65,66,69–75]. In non-hierarchical Bayesian models, information gain can be derived as a function of prediction errors and the precision of the prior and likelihood distributions, a relationship that can be approximately linear [70]. In hierarchical Bayesian inference, the update in beliefs (posterior mean changes) at each level is proportional to the precision-weighted prediction error; this update encodes the information gained from new observations [65,66,69,71,72]. Neuromodulatory arousal systems are well-situated to act as precision-weighting mechanisms in line with predictive processing frameworks [76,77]. Empirical evidence suggests that neuromodulatory systems broadcast precisionweighted prediction errors to cortical regions [11,59,66,78]. Therefore, the hypothesis that feedback-locked pupil dilation reflects a prediction error signal is similarly in line with Zenon’s main claim that pupil dilation generally reflects information gain, through precision-weighting of the prediction error. We expected a prediction error signal in pupil dilation to be proportional to the information gain.”
We have referenced previous work that has linked prediction error and information gain directly (p. 4): “The KL divergence between posterior and prior belief distributions has been previously considered to be a proxy of (precision-weighted) prediction errors [68,72].”
We have taken the following steps to remedy this error of equating “prediction error” directly with the information gain.
First, we have replaced “KL divergence” with “information gain” whenever possible throughout the manuscript for greater clarity.
Second, we have edited the section in the introduction defining information gain substantially (p. 4):
“Information gain can be operationalized within information theory as the KullbackLeibler (KL) divergence between the posterior and prior belief distributions of a Bayesian observer, representing a formalized quantity that is used to update internal models [29,79,80]. Itti and Baldi (2005)81 termed the KL divergence between posterior and prior belief distributions as “Bayesian surprise” and showed a link to the allocation of attention. The KL divergence between posterior and prior belief distributions has been previously considered to be a proxy of (precision-weighted) prediction errors[68,72]. According to Zénon’s hypothesis, if pupil dilation reflects information gain during the observation of an outcome event, such as feedback on decision accuracy, then pupil size will be expected to increase in proportion to how much novel sensory evidence is used to update current beliefs [29,63]. ”
Finally, we have made several minor textual edits to the Abstract and main text wherever possible to further clarify the proposed relationship between prediction errors and information gain.
(2) Operationalization of prediction errors based on frequency, accuracy, and their interaction:
The authors also rely on a more model-agnostic definition of the prediction error in terms of stimulus frequency ("unsigned prediction error"), accuracy, and their interaction ("signed prediction error"). While I see the point here, I would argue that this approach offers a simple approximation to the prediction error, but it is possible that factors like difficulty and effort can influence the pupil signal at the same time, which the current approach does not take into account. I recommend computing prediction errors (defined in terms of the difference between outcome and expectation) based on a simple reinforcement-learning model and analyzing the data using a pupillometry regression model in which nuisance regressors are controlled, and results are corrected for multiple comparisons.
We agree with the reviewer’s suggestion that alternatively modeling the data in a reinforcement learning paradigm would be fruitful. We adopted the ideal learner model as we were primarily focused on Information Theory, stemming from our aim to test Zenon’s hypothesis that information gain drives pupil dilation. However, we agree with the reviewer that it is worthwhile to pursue different modeling approaches in future work. We have now included a complementary linear mixed model analysis in which we controlled for the effects of the information-theoretic variables on one another, while also including the nuisance regressors of pre-feedback baseline pupil dilation and reaction times (explained in more detail below in our response to your point #4). Results including correction for multiple comparisons was reported for all pupil time course data as detailed in Methods section 2.5.
(3) The link between model-based (KL divergence) and model-agnostic (frequency- and accuracy-based) prediction errors:
I was expecting a validation analysis showing that KL divergence and model-agnostic prediction errors are correlated (in the behavioral data). This would be useful to validate the theoretical assumptions empirically.
The model limitations and the operalization of prediction error in terms of post-feedback processing do not seem to allow for a comparison of information gain and model-agnostic prediction errors in the behavioral data for the following reasons. First, the simple ideal learner model used here is not a generative model, and therefore, cannot replicate or simulate the participants responses (see also our response to your point #6 “model validation” below). Second, the behavioral dependent variables obtained are accuracy and reaction times, which both occur before feedback presentation. While accuracy and reaction times can serve as a marker of the participant’s (statistical) confidence/uncertainty following the decision interval, these behavioral measures cannot provide access to post-feedback information processing. The pupil dilation is of interest to us because the peripheral arousal system is able to provide a marker of post-feedback processing. Through the analysis presented in Figure 3, we indeed aimed to make the comparison of the model-based information gain to the model-agnostic prediction errors via the proxy variable of post-feedback pupil dilation instead of behavioral variables. To bridge the gap between the “behaviorally agnostic” model parameters and the actual performance of the participants, we examined the relationship between the model-based information gain and the post-feedback pupil dilation separately for error and correct trials as shown in Figure 3D-F & Figure 3J-L. We hope this addresses the reviewers concern and apologize in case we did not understand the reviewers suggestion here.
(4) Model-based analyses of pupil data:
I'm concerned about the authors' model-based analyses of the pupil data. The current approach is to simply compute a correlation for each model term separately (i.e., KL divergence, surprise, entropy). While the authors do show low correlations between these terms, single correlational analyses do not allow them to control for additional variables like outcome valence, prediction error (defined in terms of the difference between outcome and expectation), and additional nuisance variables like reaction time, as well as x and y coordinates of gaze.
Moreover, including entropy and KL divergence in the same regression model could, at least within each task, provide some insights into whether the pupil response to KL divergence depends on entropy. This could be achieved by including an interaction term between KL divergence and entropy in the model.
In line with the reviewer’s suggestions, we have included a complementary linear mixed model analysis in which we controlled for the effects of the information-theoretic variables on one another, while also including the nuisance regressors of pre-feedback baseline pupil dilation and reaction times. We compared the performance of two models on the post-feedback pupil dilation in each time window of interest: Modle 1 had no interaction between information gain and entropy and Model 2 included an interaction term as suggested. We did not include the x- and y- coordinates of gaze in the mixed linear model analysis, as there are multiple values of these coordinates per trial. Furthermore, regressing out the x and y- coordinates of gaze can potentially remove signal of interest in the pupil dilation data in addition to the gaze-related confounds and we did not measure absolute pupil size (Mathôt, Melmi & Castet, 2015; Hayes & Petrov, 2015). We present more sanity checks on the pre-processing pipeline as recommended by Reviewer 1.
This new analysis resulted in several additions to the Methods (see Section 2.5) and Results. In sum, we found that including an interaction term for information gain and entropy did not lead to better model fits, but sometimes lead to significantly worse fits. Overall, the results of the linear mixed model corroborated the “simple” correlation analysis across the pupil time course while accounting for the relationship to the pre-feedback baseline pupil and preceeding reaction time differences. There was only one difference to note between the correlation and linear mixed modeling analyses: for the error trials in the cue-target 2AFC task, including entropy in the model accounted for the variance previously explained by surprise.
(5) Major differences between experimental tasks:
More generally, I'm not convinced that the authors' conclusion that the pupil response to KL divergence depends on entropy is sufficiently supported by the current design. The two tasks differ on different levels (stimuli, contingencies, when learning takes place), not just in terms of entropy. In my opinion, it would be necessary to rely on a common task with two conditions that differ primarily in terms of entropy while controlling for other potentially confounding factors. I'm afraid that seemingly minor task details can dramatically change pupil responses. The positive/negative difference in the correlation with KL divergence that the authors interpret to be driven by entropy may depend on another potentially confounding factor currently not controlled.
We agree with the reviewer’s concerns and acknowledge that the speculation concerning the directional effect of entropy across trials can not be fully substantiated by the currect study. We note that Review #1 had a similar concern. Our response to Reviewer #1 addresses this concern of Reviewer #3 as well. To better align the manuscript with the above mentioned points, we have made several changes that are detailed in our response to Reviewer #1’s public review (above).
(6) Model validation:
My impression is that the ideal learner model should work well in this case. However, the authors don't directly compare model behavior to participant behavior ("posterior predictive checks") to validate the model. Therefore, it is currently unclear if the model-derived terms like KL divergence and entropy provide reasonable estimates for the participant data.
Based on our understanding, posterior predictive checks are used to assess the goodness of fit between generated (or simulated) data and observed data. Given that the “simple” ideal learner model employed in the current study is not a generative model, a posterior predictive check would not apply here (Gelman, Carlin, Stern, Dunson, Vehtari, & Rubin (2013). The ideal learner model is unable to simulate or replicate the participants’ responses and behaviors such as accuracy and reaction times; it simply computes the probability of seeing each stimulus type at each trial based on the prior distribution and the exact trial order of the stimuli presented to each participant. The model’s probabilities are computed directly from a Dirichlet distribution of values that represent the number of occurences of each stimulus-pair type for each task. The information-theoretic variables are then directly computed from these probabilities using standard formulas. The exact formulas used in the ideal learner model can be found in section 2.4.
We have now included a complementary linear mixed model analysis which also provides insight into the amount of explained variance of these information-theoretic predictors on the post-feedback pupil response, while also including the pre-feedback baseline pupil and reaction time differences (see section 3.3, Tables 3 & 4). The R<sup>2</sup> values ranged from 0.16 – 0.50 across all conditions tested.
(7) Discussion:
The authors interpret the directional effect of the pupil response w.r.t. KL divergence in terms of differences in entropy. However, I did not find a normative/computational explanation supporting this interpretation. Why should the pupil (or the central arousal system) respond differently to KL divergence depending on differences in entropy?
The current suggestion (page 24) that might go in this direction is that pupil responses are driven by uncertainty (entropy) rather than learning (quoting O'Reilly et al. (2013)). However, this might be inconsistent with the authors' overarching perspective based on Zénon (2019) stating that pupil responses reflect updating, which seems to imply learning, in my opinion. To go beyond the suggestion that the relationship between KL divergence and pupil size "needs more context" than previously assumed, I would recommend a deeper discussion of the computational underpinnings of the result.
Since we have removed the original speculative conclusion from the manuscript, we will refrain from discussing the computational underpinnings of a potential mechanism. To note as mentioned above, we have preliminary data from our own lab that contradicts our original hypothesis about the relationship between entropy and information gain on the post-feedback pupil response.
Recommendations for the authors:
Reviewer #1 (Recommendations for the authors):
Apart from the points raised in the public review above, I'd like to use the opportunity here to provide a more detailed review of potential issues, questions, and queries I have:
(1) Constriction vs. Dilation Effects:
The study observes a context-dependent relationship between KL divergence and pupil responses, where pupil dilation and constriction appear to exhibit opposing effects. However, this phenomenon raises a critical concern: Could the initial pupil constriction to visual stimuli (e.g., in the cue-target task) confound correlations with KL divergence? This potential confound warrants further clarification or control analyses to ensure that the observed effects genuinely reflect prediction error signals and are not merely a result of low-level stimulus-driven responses.
We agree with the reviewers concern and have added the following information to the limitations section in the Discussion (changes in italics below; p. 32-33).
“First, the two associative learning paradigms differed in many ways and were not directly comparable. For instance, the shape of the mean pupil response function differed across the two tasks in accordance with a visual or auditory feedback stimulus (compare Supplementary Figure 3A with Supplementary Figure 3D), and it is unclear whether these overall response differences contributed to any differences obtained between task conditions within each task. We are unable to rule out whether so-called “low level” effects such as the initial constriction to visual stimuli in the cue-target 2AFC task as compared with the dilation in response auditory stimuli in letter-color 2AFC task could confound correlations with information gain. Future work should strive to disentangle how the specific aspects of the associative learning paradigms relate to prediction errors in pupil dilation by systematically manipulating design elements within each task.”
Here, I also was curious about Supplementary Figure 1, showing 'no difference' between the two tones (indicating 'error' or 'correct'). Was this the case for FDR-corrected or uncorrected cluster statistics? Especially since the main results also showed sig. differences only for uncorrected cluster statistics (Figure 2), but were n.s. for FDR corrected. I.e. can we be sure to rule out a confound of the tones here after all?
As per the reviewer’s suggestion, we verified that there were also no significant clusters after feedback onset before applying the correction for multiple comparisons. We have added this information to Supplemenatary section 1.2 as follows:
“Results showed that the auditory tone dilated pupils on average (Supplementary Figure 1C). Crucially, however, the two tones did not differ from one another in either of the time windows of interest (Supplementary Figure 1D; no significant time points after feedback onset were obtained either before or after correcting for multiple comparisons using cluster-based permutation methods; see Section 2.5.”
Supplementary Figure 1 is showing effects cluster-corrected for multiple comparisons using cluster-based permutation tests from the MNE software package in Python (see Methods section 2.5). We have clarified that the cluster-correction was based on permutation testing in the figure legend.
(2) Participant-Specific Priors:
The ideal learner models do not account for individualised priors, assuming homogeneous learning behaviour across participants. Could incorporating participant-specific priors better reflect variability in how individuals update their beliefs during associative learning?
We have clarified in the Methods (see section 2.4) that the ideal learner models did account for participant-specific stimuli including participant-specific priors in the letter-color 2AFC task. We have added the following texts:
“We also note that while the ideal learner model for the cue-target 2AFC task used a uniform (flat) prior distribution for all participants, the model parameters were based on the participant-specific cue-target counterbalancing conditions and randomized trial order.” (p. 13)
“The prior distributions used for the letter-color 2AFC task were estimated from the randomized letter-color pairs and randomized trial order presentation in the preceding odd-ball task; this resulted in participant-specific prior distributions for the ideal learner model of the letter-color 2AFC task. The model parameters were likewise estimated from the (participant-specific) randomized trial order presented in the letter-color 2AFC task.” (p. 13)
(3) Trial-by-Trial Variability:
The analysis does not account for random effects or inter-trial variability using mixed-effects models. Including such models could provide a more robust statistical framework and ensure the observed relationships are not influenced by unaccounted participant- or trial-specific factors.
We have included a complementary linear mixed model analysis in which “subject” was modeled as a random effect on the post-feedback pupil response in each time window of interest and for each task. Across all trials, the results of the linear mixed model corroborated the “simple” correlation analysis across the pupil time course while accounting for the relationship to the prefeedback baseline pupil and preceeding reaction time differences (see section 3.3, Tables 3 & 4).
(4) Preprocessing/Analysis choices:
Before anything else, I'd like to highlight the authors' effort in providing public code (and data) in a very readable and detailed format!
We appreciate the compliment - thank you for taking the time to look at the data and code provided.
I found the idea of regressing the effect of Blinks/Saccades on the pupil trace intriguing. However, I miss a complete picture here to understand how well this actually worked, especially since it seems to be performed on already interpolated data. My main points here are:
(4.1) Why is the deconvolution performed on already interpolated data and not on 'raw' data where there are actually peaks of information to fit?
To our understanding, at least one critical reason for interpolating the data before proceeding with the deconvolution analysis is that the raw data contain many missing values (i.e., NaNs) due to the presence of blinks. Interpolating over the missing data first ensures that there are valid numerical elements in the linear algebra equations. We refer the reviewer to the methods detailed in Knapen et al. (2016) for more details on this pre-processing method.
(4.2) What is the model fit (e.g. R-squared)? If this was a poor fit for the regressors in the first place, can we trust the residuals (i.e. clean pupil trace)? Is it possible to plot the same Pupil trace of Figure 1D with a) the 'raw' pupil time-series, b) after interpolation only (both of course also mean-centered for comparison), on top of the residuals after deconvolution (already presented), so we can be sure that this is not driving the effects in a 'bad' way? I'd just like to make sure that this approach did not lead to artefacts in the residuals rather than removing them.
We thank the reviewer for this suggestion. In the Supplementary Materials, we have included a new figure (Supplementary Figure 2, copied below for convience), which illustrates the same conditions as in Figure 1D and Figure 2D, with 1) the raw data, and 2) the interpolated data before the nuisance regression. Both the raw data and interpolated data have been band-pass filtered as was done in the original pre-processing pipeline and converted to percent signal change. These figures can be compared directly to Figure 1D and Figure 2D, for the two tasks, respectively.
Of note is that the raw data seem to be dominated by responses to blinks (and/or saccades). Crucially, the pattern of results remains overall unchaged between the interpolated-only and fully pre-processed version of the data for both tasks.
In the Supplementary Materials (see Supplementary section 2), we have added the descriptives of the model fits from the deconvolution method. Model fits (R<sup>2</sup>) for the nuisance regression were generally low: cue-target 2AFC task, M = 0.03, SD = 0.02, range = [0.00, 0.07]; letter-color visual 2AFC, M = 0.08, SD = 0.04, range = [0.02, 0.16].
Furthermore, a Pearson correlation analysis between the interpolated and fully pre-processed data within the time windows of interest for both task indicated high correspondence:
Cue-target 2AFC task
Early time window: M = 0.99, SD = 0.01, range = [0.955, 1.000]
Late time window: M = 0.99, SD = 0.01, range = [0.971, 1.000]
Letter-color visual 2AFC
Early time window: M = 0.95, SD = 0.04, range = [0.803, 0.998]
Late time window: M = 0.97, SD = 0.02, range = [0.908, 0.999]
In hindsight, including the deconvolution (nuisance regression) method may not have changed the pattern of results much. However, the decision to include this deconvolution method was not data-driven; instead, it was based on the literature establishing the importance of removing variance (up to 5 s) of these blinks and saccades from cognitive effects of interest in pupil dilation (Knapen et al., 2016).
(4.3) Since this should also lead to predicted time series for the nuisance-regressors, can we see a similar effect (of what is reported for the pupil dilation) based on the blink/saccade traces of a) their predicted time series based on the deconvolution, which could indicate a problem with the interpretation of the pupil dilation effects, and b) the 'raw' blink/saccade events from the eye-tracker? I understand that this is a very exhaustive analysis so I would actually just be interested here in an averaged time-course / blink&saccade frequency of the same time-window in Figure 1D to complement the PD analysis as a sanity check.
Also included in the Supplementary Figure 2 is the data averaged as in Figure 1D and Figure 2D for the raw data and nuisance-predictor time courses (please refer to the bottom row of the sub-plots). No pattern was observed in either the raw data or the nuisance predictors as was shown in the residual time courses.
(4.4) How many samples were removed from the time series due to blinks/saccades in the first place? 150ms for both events in both directions is quite a long bit of time so I wonder how much 'original' information of the pupil was actually left in the time windows of interest that were used for subsequent interpretations.
We thank the reviewer for bringing this issue to our attention. The size of the interpolation window was based on previous literature, indicating a range of 100-200 ms as acceptable (Urai et al., 2017; Knapen et al., 2016; Winn et al., 2018). The ratio of interpolated-to-original data (across the entire trial) varied greatly between participants and between trials: cue-target 2AFC task, M = 0.262, SD = 0.242, range = [0,1]; letter-color 2AFC task, M = 0.194, SD = 0.199, range = [0,1].
We have now included a conservative analysis in which only trials with more than half (threshold = 60%) of original data are included in the analyses. Crucially, we still observe the same pattern of effects as when all data are considered across both tasks (compare the second to last row in the Supplementary Figure 2 to Figure 1D and Figure 2D).
(4.5) Was the baseline correction performed on the percentage change unit?
Yes, the baseline correction was performed on the pupil timeseries after converting to percentsignal change. We have added that information to the Methods (section 2.3).
(4.6) What metric was used to define events in the derivative as 'peaks'? I assume some sort of threshold? How was this chosen?
The threshold was chosen in a data-driven manner and was kept consistent across both tasks. The following details have been added to the Methods:
“The size of the interpolation window preceding nuisance events was based on previous literature [13,39,99]. After interpolation based on data-markers and/or missing values, remaining blinks and saccades were estimated by testing the first derivative of the pupil dilation time series against a threshold rate of change. The threshold for identifying peaks in the temporal derivative is data-driven, partially based on past work[10,14,33]. The output of each participant’s pre-processing pipeline was checked visually. Once an appropriate threshold was established at the group level, it remained the same for all participants (minimum peak height of 10 units).” (p. 8 & 11).
(5) Multicollinearity Between Variables:
Lastly, the authors state on page 13: "Furthermore, it is expected that these explanatory variables will be correlated with one another. For this reason, we did not adopt a multiple regression approach to test the relationship between the information-theoretic variables and pupil response in a single model". However, the very purpose of multiple regression is to account for and disentangle the contributions of correlated predictors, no? I might have missed something here.
We apologize for the ambiguity of our explanation in the Methods section. We originally sought to assess the overall relationship between the post-feedback response and information gain (primarily), but also surprise and entropy. Our reasoning was that these variables are often investigated in isolation across different experiments (i.e., only investigating Shannon surprise), and we would like to know what the pattern of results would look like when comparing a single information-theoretic variable to the pupil response (one-by-one). We assumed that including additional explanatory variables (that we expected to show some degree of collinearity with each other) in a regression model would affect variance attributed to them as compared with the one-on-one relationships observed with the pupil response (Morrissey & Ruxton 2018). We also acknowledge the value of a multiple regression approach on our data. Based on the suggestions by the reviewers we have included a complementary linear mixed model analysis in which we controlled for the effects of the information-theoretic variables on one another, while also including the nuisance regressors of pre-feedback baseline pupil dilation and reaction times.
This new analysis resulted in several additions to the Methods (see Section 2.5) and Results (see Tables 3 and 4). Overall, the results of the linear mixed model corroborated the “simple” correlation analysis across the pupil time course while accounting for the relationship to the prefeedback baseline pupil and preceeding reaction time differences. There was only one difference to note between the correlation and linear mixed modeling analyses: for the error trials in the cue-target 2AFC task, including entropy in the model accounted for the variance previously explained by surprise.
Reviewer #2 (Recommendations for the authors):
(1) Given the inherent temporal dependencies in pupil dynamics, characterising later pupil responses as independent of earlier ones in a three-way repeated measures ANOVA may not be appropriate. A more suitable approach might involve incorporating the earlier pupil response as a covariate in the model.
We thank the reviewer for bringing this issue to our attention. From our understanding, a repeated-measures ANOVA with factor “time window” would be appropriate in the current context for the following reasons. First, autocorrelation (closely tied to sphericity) is generally not considered a problem when only two timepoints are compared from time series data (Field, 2013; Tabachnick & Fidell, 2019). Second, the repeated-measures component of the ANOVA takes the correlated variance between time points into account in the statistical inference. Finally, as a complementary analysis, we present the results testing the interaction between the frequency and accuracy conditions across the full time courses (see Figures 1D and 2D); in these pupil time courses, any difference between the early and late time windows can be judged by the reader visually and qualitatively.
(2) Please clarify the correlations between KL divergence, surprise, entropy, and pupil response time series. Specifically, state whether these correlations account for the interrelationships between these information-theoretic measures. Given their strong correlations, partialing out these effects is crucial for accurate interpretation.
As mentioned above, based on the suggestions by the reviewers we have included a complementary linear mixed model analysis in which we controlled for the effects of the information-theoretic variables on one another, while also including the nuisance regressors of pre-feedback baseline pupil dilation and reaction times.
This new analysis resulted in several additions to the Methods (see Section 2.5) and Results (see Tables 3 and 4). Overall, the results of the linear mixed model corroborated the “simple” correlation analysis across the pupil time course while accounting for the relationship to the prefeedback baseline pupil and preceeding reaction time differences. There was only one difference to note between the correlation and linear mixed modeling analyses: for the error trials in the cue-target 2AFC task, including entropy in the model accounted for the variance previously explained by surprise.
(3) The effects observed in the late time windows appear weak (e.g., Figure 2E vs. 2F, and the generally low correlation coefficients in Figure 3). Please elaborate on the reliability and potential implications of these findings.
We have now included a complementary linear mixed model analysis which also provides insight into the amount of explained variance of these information-theoretic predictors on the post-feedback pupil response, while also including the pre-feedback baseline pupil and reaction time differences (see section 3.3, Tables 3 & 4). The R<sup>2</sup> values ranged from 0.16 – 0.50 across all conditions tested. Including the pre-feedback baseline pupil dilation as a predictor in the linear mixed model analysis consistently led to more explained variance in the post-feedback pupil response, as expected.
(4) In Figure 3 (C-J), please clarify how the trial-by-trial correlations were computed (averaged across trials or subjects). Also, specify how the standard error of the mean (SEM) was calculated (using the number of participants or trials).
The trial-by-trial correlations between the pupil signal and model parameters were computed for each participant, then the coefficients were averaged across participants for statistical inference. We have added several clarifications in the text (see section 2.5 and legends of Figure 3 and Supplementary Figure 4).
We have added “the standard error of the mean across participants” to all figure labels.
(5) For all time axes (e.g., Figure 2D), please label the ticks at 0, 0.5, 1, 1.5, 2, 2.5, and 3 seconds. Clearly indicate the duration of the feedback on the time axes. This is particularly important for interpreting the pupil dilation responses evoked by auditory feedback.
We have labeled the x-ticks every 0.5 seconds in all figures and indicated the duration of the auditory feedback in the letter-color decision task and as well as the stimuli presented in the control tasks in the Supplementary Materials.
Reviewer #3 (Recommendations for the authors):
(1) Introduction page 3: "In information theory, information gain quantifies the reduction of uncertainty about a random variable given the knowledge of another variable. In other words, information gain measures how much knowing about one variable improves the prediction or understanding of another variable."
(2) In my opinion, the description of information gain can be clarified. Currently, it is not very concrete and quite abstract. I would recommend explaining it in the context of belief updating.
We have removed these unclear statements in the Introduction. We now clearly state the following:
“Information gain can be operationalized within information theory as the KullbackLeibler (KL) divergence between the posterior and prior belief distributions of a Bayesian observer, representing a formalized quantity that is used to update internal models [29,79,80].” (p. 4)
(3) Page 4: The inconsistencies across studies are described in extreme detail. I recommend shortening this part and summarizing the inconsistencies instead of listing all of the findings separately.
As per the reviewer’s recommendation, we have shortened this part of the introduction to summarize the inconsistencies in a more concise manner as follows:
“Previous studies have shown different temporal response dynamics of prediction error signals in pupil dilation following feedback on decision outcome: While some studies suggest that the prediction error signals arise around the peak (~1 s) of the canonical impulse response function of the pupil [11,30,41,61,62,90], other studies have shown evidence that prediction error signals (also) arise considerably later with respect to feedback on choice outcome [10,25,32,41,62]. A relatively slower prediction error signal following feedback presentation may suggest deeper cognitive processing, increased cognitive load from sustained attention or ongoing uncertainty, or that the brain is integrating multiple sources of information before updating its internal model. Taken together, the literature on prediction error signals in pupil dilation following feedback on decision outcome does not converge to produce a consistent temporal signature.” (p. 5)
We would like to note some additional minor corrections to the preprint:
We have clarified the direction of the effect in Supplementary Figure 3 with the following:
“Participants who showed a larger mean difference between the 80% as compared with the 20% frequency conditions in accuracy also showed smaller differences (a larger mean difference in magnitude in the negative direction) in pupil responses between frequency conditions (see Supplementary Figure 4).”
The y-axis labels in Supplementary Figure 3 were incorrect and have been corrected as the following: “Pupil responses (80-20%)”.
We corrected typos, formatting and grammatical mistakes when discovered during the revision process. Some minor changes were made to improve clarity. Of course, we include a version of the manuscript with Tracked Changes as instructed for consideration.
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the previous reviews.
Public Reviews:
Reviewer #1 (Public Review):
Summary:
This research group has consistently performed cutting-edge research aiming to understand the role of hormones in the control of social behaviors, specifically by utilizing the genetically-tractable teleost fish, medaka, and the current work is no exception. The overall claim they make, that estrogens modulate social behaviors in males and females is supported, with important caveats. For one, there is no evidence these estrogens are generated by "neurons" as would be assumed by their main claim that it is NEUROestrogens that drive this effect. While indeed the aromatase they have investigated is expressed solely in the brain, in most teleosts, brain aromatase is only present in glial cells (astrocytes, radial glia). The authors should change this description so as not to mislead the reader. Below I detail more specific strengths and weaknesses of this manuscript.
We thank the reviewer for this positive evaluation of our work and for the helpful comments and suggestions. Regarding the concern that the term “neuroestrogens” may be misleading, we addressed this in the previous revision by consistently replacing it throughout the manuscript with “brain-derived estrogens” or “brain estrogens.”
In addition, the following sentence was added to the Introduction (line 61): “In teleost brains, including those of medaka, aromatase is exclusively localized in radial glial cells, in contrast to its neuronal localization in rodent brains (Forlano et al., 2001; Diotel et al., 2010; Takeuchi and Okubo, 2013).”
Strenghth:
Excellent use of the medaka model to disentangle the control of social behavior by sex steroid hormones
The findings are strong for the most part because deficits in the mutants are restored by the molecule (estrogens) that was no longer present due to the mutation
Presentation of the approach and findings are clear, allowing the reader to make their own inferences and compare them with the authors'
Includes multiple follow-up experiments, which leads to tests of internal replication and an impactful mechanistic proposal
Findings are provocative not just for teleost researchers, but for other species since, as the authors point out, the data suggest mechanisms of estrogenic control of social behaviors may be evolutionary ancient
We thank the reviewer again for their positive evaluation of our work.
Weakness:
As stated in the summary, the authors are attributing the estrogen source to neurons and there isn't evidence this is the case. The impact of the findings doesn't rest on this either
As mentioned above, we addressed this in the previous revision by replacing “neuroestrogens” with “brain-derived estrogens” or “brain estrogens” throughout the manuscript. In addition, the following sentence was added to the Introduction (line 61): “In teleost brains, including those of medaka, aromatase is exclusively localized in radial glial cells, in contrast to its neuronal localization in rodent brains (Forlano et al., 2001; Diotel et al., 2010; Takeuchi and Okubo, 2013).”
The d4 versus d8 esr2a mutants showed different results for aggression. The meaning and implications of this finding are not discussed, leaving the reader wondering
This comment is the same as one raised in the first review (Reviewer #1’s comment 2 on weaknesses), which we already addressed in our initial revision. For the reviewer’s convenience, we provide the response below:
Line 300: As the reviewer correctly noted, circles were significantly reduced in mutant males of the Δ8 line, whereas no significant reduction was observed in those of the Δ4 line. However, a tendency toward reduction was evident in the Δ4 line (P = 0.1512), and both lines showed significant differences in fin displays. Based on these findings, we believe our conclusion that esr2a<sup>−/−</sup> males exhibit reduced aggression remains valid. To clarify this point and address potential reader concerns, we have revised the text as follows: “esr2a<sup>−/−</sup> males exhibited significantly fewer fin displays (P = 0.0461 and 0.0293 for Δ8 and Δ4 lines, respectively) and circles (P = 0.0446 and 0.1512 for Δ8 and Δ4 lines, respectively) than their wild-type siblings (Fig. 5L; Fig. S8E), suggesting less aggression” was edited to read “esr2a<sup>−/−</sup> males from both the Δ8 and Δ4 lines exhibited significantly fewer fin displays than their wild-type siblings (P = 0.0461 and 0.0293, respectively). Circles followed a similar pattern, with a significant reduction in the Δ8 line (P = 0.0446) and a comparable but non-significant decrease in the Δ4 line (P =0.1512) (Figure 5L, Figure 5—figure supplement 3E), showing less aggression.”
Lack of attribution of previous published work from other research groups that would provide the proper context of the present study
This comment is also the same as one raised in the first review (Reviewer #1’s comment 3 on weaknesses). In our previous revision, in response to this comment, we cited the relevant references (Hallgren et al., 2006; O’Connell and Hofmann, 2012; Huffman et al., 2013; Jalabert et al., 2015; Yong et al., 2017; Alward et al., 2020; Ogino et al., 2023) in the appropriate sections. We also added the following new references and revised the Introduction and Discussion accordingly:
(2) Alward BA, Laud VA, Skalnik CJ, York RA, Juntti SA, Fernald RD. 2020. Modular genetic control of social status in a cichlid fish. Proceedings of the National Academy of Sciences of the United States of America 117:28167–28174. DOI: https://doi.org/10.1073/pnas.2008925117
(39) O’Connell LA, Hofmann HA. 2012. Social status predicts how sex steroid receptors regulate complex behavior across levels of biological organization. Endocrinology 153:1341–1351. DOI:https://doi.org/10.1210/en.2011-1663
(54) Yong L, Thet Z, Zhu Y. 2017. Genetic editing of the androgen receptor contributes to impaired male courtship behavior in zebrafish. Journal of Experimental Biology 220:3017–3021.DOI:https://doi.org/10.1242/jeb.161596
There are a surprising number of citations not included; some of the ones not included argue against the authors' claims that their findings were "contrary to expectation"
In our previous revision, we cited the relevant references (Hallgren et al., 2006; O’Connell and Hofmann, 2012; Huffman et al., 2013; Jalabert et al., 2015) in the Introduction. We also revised the text to remove phrases such as “contrary to expectation” and “unexpected.”
The experimental design for studying aggression in males has flaws. A standard test like a residentintruder test should be used.
Following this comment, we have attempted additional aggression assays using the resident-intruder paradigm. However, these experiments did not produce consistent or interpretable results. As noted in our previous revision, medaka naturally form shoals and exhibit weak territoriality, and even slight differences in dominance between a resident and an intruder can markedly increase variability, reducing data reliability. Therefore, we believe that the approach used in the present study provides a more suitable assessment of aggression in medaka, regardless of territorial tendencies. We will continue to explore potential refinements in future studies and respectfully ask the reviewer to evaluate the present work based on the assay used here.
While they investigate males and females, there are fewer experiments and explanations for the female results, making it feel like a small addition or an aside
While we did not adopt this comment in our previous revision, we have carefully reconsidered the reviewers’ feedback and have now decided to remove the female data. This change allows us to present a more focused and cohesive story centered on males. The specific revisions are outlined below:
Abstract
Line 25: The text “, thereby revealing a previously unappreciated mode of action of brain-derived estrogens. We additionally show that female fish lacking Cyp19a1b are less receptive to male courtship and conversely court other females, highlighting the significance of brain-derived estrogens in establishing sex-typical behaviors in both sexes.” has been revised to “. Taken together, these findings reveal a previously unappreciated mode of action of brain-derived estrogens in shaping male-typical behaviors.”
Results
Line 88: The text “Loss of cyp19a1b function in these fish was verified by measuring brain and peripheral levels of sex steroids. As expected, brain estradiol-17β (E2) in both male and female homozygous mutants (cyp19a1b<sup>−/−</sup>) was significantly reduced to 16% and 50%, respectively, of the levels in their wild-type (cyp19a1b<sup>+/+</sup>) siblings (P = 0.0037, males; P = 0.0092, females) (Fig. 1, A and B). In males, brain E2 in heterozygotes (cyp19a1b<sup>−/−</sup>) was also reduced to 45% of the level in wild-type siblings (P = 0.0284) (Fig. 1A), indicating a dosage effect of cyp19a1b mutation. In contrast, peripheral E2 levels were unaltered in both cyp19a1b<sup>−/−</sup> males and females (Fig. S1, C and D), consistent with the expected functioning of Cyp19a1b primarily in the brain. Strikingly, brain levels of testosterone, as opposed to E2, increased 2.2-fold in cyp19a1b<sup>−/−</sup> males relative to wild-type siblings (P = 0.0006) (Fig. 1A). Similarly, brain 11KT levels in cyp19a1b<sup>−/−</sup> males and females increased 6.2- and 1.9-fold, respectively, versus wild-type siblings (P = 0.0007, males; P = 0.0316, females) (Fig. 1, A and B). These results show that cyp19a1b-deficient fish have reduced estrogen levels coupled with increased androgen levels in the brain, confirming the loss of cyp19a1b function. They also suggest that the majority of estrogens in the male brain and half of those in the female brain are synthesized locally in the brain. In addition, peripheral 11KT levels in cyp19a1b<sup>−/−</sup> males and females increased 3.7- and 1.8-fold, respectively (P = 0.0789, males; P = 0.0118, females) (Fig. S1, C and D), indicating peripheral influence in addition to central effects.” has been revised to “Loss of cyp19a1b function in these fish was verified by measuring brain and peripheral levels of sex steroids in males. As expected, brain estradiol-17β (E2) in homozygous mutants (cyp19a1b<sup>−/−</sup>) was significantly reduced to 16% of the levels in wild-type (cyp19a1b<sup>+/+</sup>) siblings (P = 0.0037) (Figure 1A). Brain E2 in heterozygotes (cyp19a1b<sup>+/−</sup>) was also reduced to 45% of wild-type levels (P = 0.0284) (Figure 1A), indicating a dosage effect of the cyp19a1b mutation. In contrast, peripheral E2 levels were unaltered in cyp19a1b<sup>−/−</sup> males (Figure 1B), consistent with the expected functioning of Cyp19a1b primarily in the brain. Strikingly, brain testosterone levels, as opposed to E2, increased 2.2-fold in cyp19a1b<sup>−/−</sup> males relative to wild-type siblings (P = 0.0006) (Figure 1A). Similarly, brain 11KT levels increased 6.2-fold (P = 0.0007) (Figure 1A). These results indicate that cyp19a1b-deficient males have reduced estrogen coupled with elevated androgen levels in the brain, confirming the loss of cyp19a1b function. They also suggest that the majority of estrogens in the male brain are synthesized locally in the brain. Peripheral 11KT levels also increased 3.7-fold in cyp19a1b<sup>−/−</sup> males (P = 0.0789) (Figure 1B), indicating peripheral influence in addition to central effects.”
Line 211: “expression of vt in the pNVT of cyp19a1b<sup>−/−</sup> males was significantly reduced to 18% as compared with cyp19a1b<sup>+/+</sup> males (P = 0.0040), a level comparable to that observed in females” has been revised to “expression of vt in the pNVT of cyp19a1b<sup>−/−</sup> males was significantly reduced to 18% as compared with cyp19a1b<sup>+/+</sup> males (P = 0.0040).”
The subsection entitled “cyp19a1b-deficient females are less receptive to males and instead court other females,” which followed line 311, has been removed.
Discussion
The two paragraphs between lines 373 and 374, which addressed the female data, have been removed.
Materials and methods
Line 433: “males and females” has been changed to “males”.
Line 457: “focal fish” has been changed to “focal male”.
Line 458: “stimulus fish” has been changed to “stimulus female”.
Line 458: “Fig. 6, E and F, ” has been deleted.
Line 460: “; wild-type males in Fig. 6, A to C” has been deleted.
Line 466: The text “The period of interaction/recording was extended to 2 hours in tests of courtship displays received from the stimulus esr2b-deficient female and in tests of mating behavior between females, because they take longer to initiate courtship (12). In tests using an esr2b-deficient female as the stimulus fish, where the latency to spawn could not be calculated because these fish were unreceptive to males and did not spawn, the sexual motivation of the focal fish was instead assessed by counting the number of courtship displays and wrapping attempts in 30 min. The number of these mating acts was also counted in tests to evaluate the receptivity of females. In tests of mating behavior between two females, the stimulus female was marked with a small notch in the caudal fin to distinguish it from the focal female.” has been revised to “In tests using an esr2b-deficient female as the stimulus fish, the latency to spawn could not be calculated because the female was unreceptive to males and did not spawn. Therefore, the sexual motivation of the focal male was assessed by counting the number of courtship displays and wrapping attempts in 30 min. To evaluate courtship displays performed by stimulus esr2bdeficient females toward focal males, the recording period was extended to 2 hours, as these females take longer to initiate courtship (Nishiike et al., 2021). In all video analyses, the researcher was blind to the fish genotype and treatment.”
Line 499: “brains dissected from males and females of the cyp19a1b-deficient line (analysis of ara, arb, vt, gal, npba, and esr2b) and males of the esr1-, esr2a-, and esr2b-deficient lines” has been revised to “male brains from the cyp19a1b-deficient line (analysis of ara, arb, vt, and gal) and from the esr1-, esr2a-, and esr2b-deficient lines.”
Line 504: “After color development for 15 min (gal), 40 min (npba), 2 hours (vt), or overnight (ara, arb, and esr2b)” has been revised to “After color development for 15 min (gal), 2 hours (vt), or overnight (ara and arb).”
Line 516: “Thermo Fisher Scientific, Waltham, MA” has been changed to “Thermo Fisher Scientific” to avoid redundancy.
Line 565: The subsection entitled “Measurement of spatial distances between fish” has been removed.
Line 585: “6/10 cyp19a1b<sup>+/+</sup>, 3/10 cyp19a1b<sup>+/−</sup>, and 6/10 cyp19a1b<sup>−/−</sup> females were excluded in Fig. 6B;” has been deleted.
References
The following references have been removed:
Capel B. 2017. Vertebrate sex determination: evolutionary plasticity of a fundamental switch. Nature Reviews Genetics 18:675–689. DOI: https://doi.org/10.1038/nrg.2017.60
Hiraki T, Nakasone K, Hosono K, Kawabata Y, Nagahama Y, Okubo K. 2014. Neuropeptide B is femalespecifically expressed in the telencephalic and preoptic nuclei of the medaka brain. Endocrinology 155:1021–1032. DOI: https://doi.org/10.1210/en.2013-1806
Juntti SA, Hilliard AT, Kent KR, Kumar A, Nguyen A, Jimenez MA, Loveland JL, Mourrain P, Fernald RD. 2016. A neural basis for control of cichlid female reproductive behavior by prostaglandin F2α. Current Biology 26:943–949. DOI: https://doi.org/10.1016/j.cub.2016.01.067
Kimchi T, Xu J, Dulac C. 2007. A functional circuit underlying male sexual behaviour in the female mouse brain. Nature 448:1009–1014. DOI: https://doi.org/10.1038/nature06089
Kobayashi M, Stacey N. 1993. Prostaglandin-induced female spawning behavior in goldfish (Carassius auratus) appears independent of ovarian influence. Hormones and Behavior 27:38–55.
DOI:https://doi.org/10.1006/hbeh.1993.1004
Liu H, Todd EV, Lokman PM, Lamm MS, Godwin JR, Gemmell NJ. 2017. Sexual plasticity: a fishy tale. Molecular Reproduction and Development 84:171–194. DOI: https://doi.org/10.1002/mrd.22691
Munakata A, Kobayashi M. 2010. Endocrine control of sexual behavior in teleost fish. General and Comparative Endocrinology 165:456–468. DOI: https://doi.org/10.1016/j.ygcen.2009.04.011
Nugent BM, Wright CL, Shetty AC, Hodes GE, Lenz KM, Mahurkar A, Russo SJ, Devine SE, McCarthy MM. 2015. Brain feminization requires active repression of masculinization via DNA methylation. Nature Neuroscience 18:690–697. DOI: https://doi.org/10.1038/nn.3988
Shaw K, Therrien M, Lu C, Liu X, Trudeau VL. 2023. Mutation of brain aromatase disrupts spawning behavior and reproductive health in female zebrafish. Frontiers in Endocrinology 14:1225199.
DOI:https://doi.org/10.3389/fendo.2023.1225199
Stacey NE. 1976. Effects of indomethacin and prostaglandins on the spawning behaviour of female goldfish. Prostaglandins 12:113–126. DOI: https://doi.org/10.1016/s0090-6980(76)80010-x
Figure 1
Panel B, which originally showed steroid levels in female brains, has been replaced with steroid levels in the periphery of males, originally presented in Figure S1, panel C. Accordingly, the legend “(A and B) Levels of E2, testosterone, and 11KT in the brain of adult cyp19a1b<sup>+/+</sup>, cyp19a1b<sup>+/−</sup>, and cyp19a1b<sup>−/−</sup> males (A) and females (B) (n = 3 per genotype and sex).” has been revised to “(A, B) Levels of E2, testosterone, and 11KT in the brain (A) and periphery (B) of adult cyp19a1b<sup>+/+</sup>, cyp19a1b<sup>+/−</sup>, and cyp19a1b<sup>−/−</sup> males (n = 3 per genotype).”
Figure 3
The female data have been deleted from Figure 3. The revised Figure 3 is presented.
The corresponding legend text has been revised as follows:
Line 862: “males and females (n = 4 and 5 per genotype for males and females, respectively)” has been changed to “males (n = 4 per genotype)”.
Line 864: “males and females (n = 4 except for cyp19a1b<sup>+/+</sup> males, where n = 3)” has been changed to “males (n = 3 and 4, respectively)”.
Figure 6
Figure 6 and its legend have been removed.
Figure 1—figure supplement 1
Panel C, showing male data, has been moved to Figure 1B, as described above, while panel D, showing female data, has been deleted. The corresponding legend “(C and D) Levels of E2, testosterone, and 11KT in the periphery of adult cyp19a1b<sup>+/+</sup>, cyp19a1b<sup>+/−</sup>, and cyp19a1b<sup>−/−</sup> males (C) and females (D) (n = 3 per genotype and sex). Statistical differences were assessed by Bonferroni’s post hoc test (C and D). Error bars represent SEM. *P < 0.05.” has also been removed.
Line 804: Following this change, the figure title has been updated from “Generation of cyp19a1bdeficient medaka and evaluation of peripheral sex steroid levels” to “Generation of cyp19a1b-deficient medaka.”
The statistics comparing "experimental to experimental" and "control to experimental" isn't appropriate
This comment is the same as one raised in the first review (Reviewer #1’s comment 7 on weaknesses), which we already addressed in our initial revision. For the reviewer’s convenience, we provide the response below:
The reviewer raised concerns about the statistical analysis used for Figures 4C and 4E, suggesting that Bonferroni’s test should be used instead of Dunnett’s test. However, Dunnett’s test is commonly used to compare treatment groups to a reference group that receives no treatment, as in our study. Since we do not compare the treated groups with each other, we believe Dunnett’s test is the most appropriate choice.
Line 576: The reviewer’s concern may have arisen from the phrase “comparisons between control and experimental groups” in the Materials and methods. We have revised it to “comparisons between untreated and E2-treated groups in Figure 4C and D” for clarity.
Reviewer #3 (Public Review):
Summary:
Taking advantage of the existence in fish of two genes coding for estrogen synthase, the enzyme aromatase, one mostly expressed in the brain (Cyp19a1b) and the other mostly found in the gonads (Cyp19a1a), this study investigates the role of brain-derived estrogens in the control of sexual and aggressive behavior in medaka. The constitutive deletion of Cyp19a1b markedly reduced brain estrogen content in males and to a lesser extent in females. These effects are accompanied by reduced sexual and aggressive behavior in males and reduced preference for males in females. These effects are reversed by adult treatment with supporting a role for estrogens. The deletion of Cyp19a1b is associated with a reduced expression of the genes coding for the two androgen receptors, ara and arb, in brain regions involved in the regulation of social behavior. The analysis of the gene expression and behavior of mutants of estrogen receptors indicates that these effects are likely mediated by the activation of the esr1 and esr2a isoforms. These results provide valuable insight into the role of estrogens in social behavior in the most abundant vertebrate taxon, however the conclusion of brain-derived estrogens awaits definitive confirmation.
We thank this reviewer for their positive evaluation of our work and comments that have improved the manuscript.
Strength:
Evaluation of the role of brain "specific" Cyp19a1 in male teleost fish, which as a taxon are more abundant and yet proportionally less studied that the most common birds and rodents. Therefore, evaluating the generalizability of results from higher vertebrates is important. This approach also offers great potential to study the role of brain estrogen production in females, an understudied question in all taxa.
Results obtained from multiple mutant lines converge to show that estrogen signaling, likely synthesized in the brain drives aspects of male sexual behavior.
The comparative discussion of the age-dependent abundance of brain aromatase in fish vs mammals and its role in organization vs activation is important beyond the study of the targeted species. - The authors have made important corrections to tone down some of the conclusions which are more in line with the results.
We thank the reviewer again for their positive evaluation of our work and the revisions we have made.
weaknesses:
No evaluation of the mRNA and protein products of Cyp19a1b and ESR2a are presented, such that there is no proper demonstration that the mutation indeed leads to aromatase reduction. The conclusion that these effects dependent on brain derived estrogens is therefore only supported by measures of E2 with an EIA kit that is not validated. No discussion of these shortcomings is provided in the discussion thus further weakening the conclusion manuscript.
In response to this and other comments, we have now provided direct validation that the cyp19a1b mutation in our medaka leads to loss of function. Real-time PCR analysis showed that cyp19a1b transcript levels in the brain were reduced by approximately half in cyp19a1b<sup>+/−</sup> males and were nearly absent in cyp19a1b<sup>−/−</sup> males, consistent with nonsense-mediated mRNA decay
In addition, AlphaFold 3-based structural modeling indicated that the mutant Cyp19a1b protein lacks essential motifs, including the aromatic region and heme-binding loop, and exhibits severe conformational distortion (see figure; key structural features are annotated as follows: membrane helix (blue), aromatic region (red), and heme-binding loop (orange)).
Results:
Line 101: The following text has been added: “Loss of cyp19a1b function was further confirmed by measuring cyp19a1b transcript levels in the brain and by predicting the three-dimensional structure of the mutant protein. Real-time PCR revealed that transcript levels were reduced by half in cyp19a1b<sup>+/−</sup> males and were nearly undetectable in cyp19a1b<sup>−/−</sup> males, presumably as a result of nonsense-mediated mRNA decay (Lindeboom et al., 2019) (Figure 1C). The wild-type protein, modeled by AlphaFold 3, exhibited a typical cytochrome P450 fold, including the membrane helix, aromatic region, and hemebinding loop, all arranged in the expected configuration (Figure 1—figure supplement 1C). The mutant protein, in contrast, was severely truncated, retaining only the membrane helix (Figure 1—figure supplement 1C). The absence of essential domains strongly indicates that the allele encodes a nonfunctional Cyp19a1b protein. Together, transcript and structural analyses consistently demonstrate that the mutation generated in this study causes a complete loss of cyp19a1b function.”
Materials and methods
Line 438: A subsection entitled “Real-time PCR” has been added. The text of this subsection is as follows: “Total RNA was isolated from the brains of cyp19a1b<sup>+/+</sup>, cyp19a1b<sup>+/−</sup>, and cyp19a1b<sup>−/−</sup> males using the RNeasy Plus Universal Mini Kit (Qiagen, Hilden, Germany). cDNA was synthesized with the SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA). Real-time PCR was performed on the LightCycler 480 System II using the LightCycler 480 SYBR Green I Master (Roche Diagnostics). Melting curve analysis was conducted to verify that a single amplicon was obtained in each sample. The β-actin gene (actb; GenBank accession number NM_001104808) was used to normalize the levels of target transcripts. The primers used for real-time PCR are shown in Supplementary file 2.”
Line 448: A subsection entitled “Protein structure prediction” has been added. The text of this subsection is as follows: “Structural predictions of Cyp19a1b proteins were conducted using AlphaFold 3 (Abramson et al., 2024). Amino acid sequences corresponding to the wild-type allele and the mutant allele generated in this study were submitted to the AlphaFold 3 prediction server. The resulting models were visualized with PyMOL (Schrödinger, New York, NY), and key structural features, including the membrane helix, aromatic region, and heme-binding loop, were annotated.”
References
The following two references have been added:
Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, Ronneberger O, Willmore L, Ballard AJ, Bambrick J, Bodenstein SW, Evans DA, Hung CC, O'Neill M, Reiman D, Tunyasuvunakool K, Wu Z, Žemgulytė A, Arvaniti E, Beattie C, Bertolli O, Bridgland A, Cherepanov A, Congreve M, CowenRivers AI, Cowie A, Figurnov M, Fuchs FB, Gladman H, Jain R, Khan YA, Low CMR, Perlin K, Potapenko A, Savy P, Singh S, Stecula A, Thillaisundaram A, Tong C, Yakneen S, Zhong ED, Zielinski M, Žídek A, Bapst V, Kohli P, Jaderberg M, Hassabis D, Jumper JM. 2024. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630:493–500. DOI: https://doi.org/10.1038/s41586-024-07487-w
Lindeboom RGH, Vermeulen M, Lehner B, Supek F. 2019. The impact of nonsense-mediated mRNA decay on genetic disease, gene editing and cancer immunotherapy. Nature Genetics 51:1645–1651.DOI:https://doi.org/10.1038/s41588-019-0517-5
Figure 1
The real-time PCR results described above have been incorporated in Figure 1, panel C, with the corresponding legend provided below (line 788).
(C) Brain cyp19a1b transcript levels in cyp19a1b<sup>+/+</sup>, cyp19a1b<sup>+/−</sup>, and cyp19a1b<sup>−/−</sup> males (n = 6 per genotype). Mean value for cyp19a1b<sup>+/+</sup> males was arbitrarily set to 1.
The subsequent panels have been renumbered accordingly. The entirety of the revised Figure 1.
Figure 1—figure supplement 1
The AlphaFold 3-generated structural models described above have been incorporated in Figure 1— figure supplement 1, panel C, with the corresponding legend provided below (line 811).
(C) Predicted three-dimensional structures of wild-type (left) and mutant (right) Cyp19a1b proteins. Key structural features are annotated as follows: membrane helix (blue), aromatic region (red), and heme-binding loop (orange).
The entirety of the revised Figure 1—figure supplement 1 is presented
The information on the primers used for real-time PCR has been included in Supplementary file 2.
The functional deficiency of esr2a was already addressed in the previous revision. For clarity, we have reproduced the relevant information here.
A previous study reported that female medaka lacking esr2a fail to release eggs due to oviduct atresia (Kayo et al., 2019, Sci Rep 9:8868). Similarly, in this study, some esr2a-deficient females exhibited spawning behavior but were unable to release eggs, although the sample size was limited (Δ8 line: 2/3; Δ4 line: 1/1). In contrast, this was not observed in wild-type females (Δ8 line: 0/12; Δ4 line: 0/11). These results support the effective loss of esr2a function. To incorporate this information into the manuscript, the following text has been added to the Materials and methods (line 423): “A previous study reported that esr2a-deficient female medaka cannot release eggs due to oviduct atresia (Kayo et al., 2019). Likewise, some esr2a-deficient females generated in this study, despite the limited sample size, exhibited spawning behavior but were unable to release eggs (Δ8 line: 2/3; Δ4 line: 1/1), while such failure was not observed in wild-type females (Δ8 line: 0/12; Δ4 line: 0/11). These results support the effective loss of esr2a function.”
Most experiments are weakly powered (low sample size).
This comment is essentially the same as one raised in the first review (Reviewer #3’s comment 7 on weaknesses). We acknowledge the reviewer’s concern that the histological analyses were weakly powered due to the limited sample size. In our earlier revision, we responded as follows:
Histological analyses were conducted with a relatively small sample size, as our previous experience suggested that interindividual variability in the results would not be substantial. Since significant differences were detected in many analyses, further increasing the sample size was deemed unnecessary.
The variability of the mRNA content for a same target gene between experiments (genotype comparison vs E2 treatment comparison) raises questions about the reproducibility of the data (apparent disappearance of genotype effect).
This comment is the same as one raised in the first review (Reviewer #3’s comment 8 on weaknesses), which we already addressed in our initial revision. For the reviewer’s convenience, we provide the response below:
As the reviewer pointed out, the overall area of ara expression is larger in Figure 2J than in Figure 2F. However, the relative area ratios of ara expression among brain nuclei are consistent between the two figures, indicating the reproducibility of the results. Thus, this difference is unlikely to affect the conclusions of this study.
Additionally, the differences in ara expression in pPPp and arb expression in aPPp between wild-type and cyp19a1b-deficient males appear less pronounced in Figures 2J and 2K than in Figures 2F and 2H. This is likely attributable to the smaller sample size used in the experiments for Figures 2J and 2K, resulting in less distinct differences. However, as the same genotype-dependent trends are observed in both sets of figures, the conclusion that ara and arb expression is reduced in cyp19a1b-deficient male brains remains valid.
Conclusions:
Overall, the claims regarding role of estrogens originating in the brain on male sexual behavior is supported by converging evidence from multiple mutant lines. The role of brain-derived estrogens on gene expression in the brain is weaker as are the results in females.
We appreciate the reviewer’s positive evaluation of our findings on male behavior. The concern regarding the role of brain-derived estrogens in gene expression has been addressed in our rebuttal, and the female data have been removed so that the analysis now focuses on males. The specific revisions for removing the female data are described in Response to reviewer #1’s comment 6 on weaknesses.
Recommendations For The Authors:
Reviewer #1 (Recommendations For The Authors):
The manuscript is improved slightly. I am thankful the authors addressed some concerns, but for several concerns the referees raised, the authors acknowledged them yet did not make corresponding changes to the manuscript or disagreed that they were issues at all without explanation. All reviewers had issues with the imbalanced focus on males versus females and the male aggression assay. Yet, they did not perform additional experiments or even make changes to the framing and scope of the manuscript. If the authors had removed the female data, they may have had a more cohesive story, but then they would still be left with inadequate behavior assays in the males. If the authors don't have the time or resources to perform the additional work, then they should have said so. However, the work would be incomplete relative to the claims. That is a key point here. If they change their scope and claims, the authors avoid overstating their findings. I want to see this work published because I believe it moves the field forward. But the authors need to be realistic in their interpretations of their data.
In response to this and related comments, we have removed the female data and focused the manuscript on analyses in males. The specific revisions are described in Response to reviewer #1’s comment 6 on weaknesses. Additionally, we have validated that the cyp19a1b mutation in our medaka leads to loss of function (see Response to reviewer #3’s comment 1 on weaknesses), which further strengthens the reliability of our conclusions regarding male behavior.
I agree with the reviewer who said we need to see validation of the absence of functional cyp19a1 b in the brain. However, the results from staining for the protein and performing in situ could be quizzical. Indeed, there aren't antibodies that could distinguish between aromatase a and b, and it is not uncommon for expression of a mutated gene to be normal. One approach they could do is measure aromatase activity, but they are *sort of* doing that by measuring brain E2. It's not perfect, but we teleost folks are limited in these areas. At the very least, they should show the predicted protein structure of the mutated aromatase alleles. It could show clearly that the tertiary structure is utterly absent, giving more support to the fact that their aromatase gene is non-functional.
As noted above, we have further validated the loss of cyp19a1b function by measuring cyp19a1b transcript levels in the brain and predicting the three-dimensional structure of the mutant protein. These analyses confirmed that cyp19a1b function is indeed lost, thereby increasing the reliability of our conclusions. For further details, please refer to Response to reviewer #3’s comment 1 on weaknesses.
With all of this said, the work is important, and it is possible that with a reframing of the impact of their work in the context of their findings, I could consider the work complete. I think with a proper reframing, the work is still impactful.
In accordance with this feedback, and as described above, we have reframed the manuscript by removing the female data and focusing exclusively on males. This revision clarifies the scope of our study and reinforces the support for our conclusions. For further details, please refer to Response to reviewer #1’s comment 6 on weaknesses.
(1) Clearly state in the Figure 1 legend that each data point for male aggressive behaviors represents the total # of behaviors calculated over the 4 males in each experimental tank.
In response to this comment, we have revised the legend of Figure 1K (line 797). The original legend, “(K) Total number of each aggressive act observed among cyp19a1b<sup>+/+</sup>, cyp19a1b<sup>+/−</sup>, or cyp19a1<sup>−/−</sup> males in the tank (n = 6, 7, and 5, respectively),” has been updated to “(K) Total number of each aggressive act performed by cyp19a1b<sup>+/+</sup>, cyp19a1b<sup>+/−</sup>, and cyp19a1b<sup>−/−</sup> males. Each data point represents the sum of acts recorded for the 4 males of the same genotype in a single tank (n = 6, 7, and 5 tanks, respectively).” This clarifies that each data point reflects the total behaviors of the 4 males within each tank.
(2) The authors wrote under "Response to reviewer #1's major comment "...the development of male behaviors may require moderate neuroestrogen levels that are sufficient to induce the expression of ara and arb, but not esr2b, in the underlying neural circuitry": "This may account for the lack of aggression recovery in E2-treated cyp19a1b-deficient males in this study.".
What is meant by the latter statement? What accounts for the lack of aggression? The lack of increase in esr2b? Please clarify.
Line 365: In response to this comment, “This may account for the lack of aggression recovery in E2treated cyp19a1b-deficient males in this study.” has been revised to “Considering this, the lack of aggression recovery in E2-treated cyp19a1b-deficient males in this study may be explained by the possibility that the E2 dose used was sufficient to induce not only ara and arb but also esr2b expression in aggression-relevant circuits, which potentially suppressed aggression.”
This revision clarifies that, while moderate brain estrogen levels are sufficient to promote male behaviors via induction of ara and arb, the E2 dose used in this study may have additionally induced esr2b in circuits relevant to aggression, potentially underlying the lack of aggression recovery.
(3) This is a continuation of my comment/concern directly above. If the induction of ara and arb aren't enough, then how can, as the authors state, androgen signaling be the primary driver of these behaviors?
In response to this follow-up comment, we would like to clarify that, as described above, the lack of aggression recovery in E2-treated cyp19a1b-deficient males is not due to insufficient induction of ara and arb, but instead is likely because esr2b was also induced in aggression-relevant circuits, which may have suppressed aggression. Therefore, the concern that androgen signaling cannot be the primary driver of these behaviors is not applicable.
(4) The authors' point about sticking with the terminology for the ar genes as "ara" and "arb" is not convincing. The whole point of needing a change to match the field of neuroendocrinology as a whole (that is, across all vertebrates) is researchers, especially those with high standing like the Okubo group, adopt the new terminology. Indeed, the Okubo group is THE leader in medaka neuroendocrinology. It would go a long way if they began adopting the new terminology of "ar1" and "ar2". I understand this may be laborious to a degree, and each group can choose to use their terminology, but I'd be remiss if I didn't express my opinion that changing the terminology could help our field as a whole.
We sincerely appreciate the reviewer’s thoughtful comments regarding nomenclature consistency in vertebrate neuroendocrinology. We understand the motivation behind the suggestion to adopt ar1 and ar2. However, we consider the established nomenclature of ara and arb to be more appropriate for the following reasons.
First, adopting the ar1/ar2 nomenclature would introduce a discrepancy between gene and protein symbols. According to the NCBI International Protein Nomenclature Guidelines (Section 2B.Abbreviations and symbols;
https://www.ncbi.nlm.nih.gov/genbank/internatprot_nomenguide/), the ZFIN Zebrafish Nomenclature Conventions (Section 2. PROTEINS:https://zfin.atlassian.net/wiki/spaces/general/pages/1818394635/ZFIN+Zebrafish+Nomenclature+Con ventions), and the author guidelines of many journal
(e.g.,https://academic.oup.com/molehr/pages/Gene_And_Protein_Nomenclature), gene and protein symbols should be identical (with proteins designated in non-italic font and with the first letter capitalized). Maintaining consistency between gene and protein symbols helps avoid unnecessary confusion. The ara/arb nomenclature allows this, whereas ar1/ar2 does not.
Second, the two androgen receptor genes in teleosts are paralogs derived from the third round of wholegenome duplication that occurred early in teleost evolution. For such duplicated genes, the ZFIN Zebrafish Nomenclature Conventions (Section 1.2. Duplicated genes) recommend appending the suffixes “a” and “b” to the approved symbol of the human or mouse ortholog. This convention clearly indicates that these genes are whole-genome duplication paralogs and provides an intuitive way to represent orthologous and paralogous relationships between teleost genes and those of other vertebrates. As a result, it has been widely adopted, and we consider it logical and beneficial to apply the same principle to androgen receptors.
In light of these considerations, we respectfully maintain that the ara/arb nomenclature is more suitable for the present manuscript than the alternative ar1/ar2 system.
(5) In the discussion please discuss these potentially unexpected findings.
(a) gal was unaffected in female cyp19a1 mutants, but they exhibit mating behaviors towards females. Given gal is higher in males and these females act like females, what does this mean about the function of gal/its utility in being a male-specific marker (is it one??)?
(b) esr2b expression is higher in female cyp19a1 mutants. this is unexpected as well given esr2b is required for female-typical mating and is higher in females compared to males and E2 increases esr2b expression. please explain...well, what this means for our idea of what esr2b expression tell us.
We thank the reviewer for the insightful comments. As the female data have been removed from the manuscript, discussion of these findings in female cyp19a1b mutants is no longer necessary.
Reviewer #3 (Recommendations For The Authors):
The authors have addressed a number of answers to the reviewer's comments, notably they provided missing methodological information and rephrased the text. However, the authors have not addressed the main issues raised by the reviewers. Notably, it is regrettable that the reduced amount of brain aromatase cannot be confirmed, this seems to be the primary step when validating a new mutant. Even if protein products of the two genes may not be discriminated (which I can understand), it should be possible to evaluate the expression of a common messenger and/or peptide and confirm that aromatase expression is reduced in the brain. Since Cyp19a1b is relatively more abundant in the brain Cyp19a1a, this would strengthen the conclusion and provide confidence that the mutant indeed does silence aromatase expression in the brain. Although these short comings are acknowledged in the rebuttal letter, this is not mentioned in the discussion. Doing so would make the manuscript more transparent and clearer.
As noted in Response to reviewer #3’s comment 1 on weaknesses, we have validated the loss of Cyp19a1b function by measuring its transcript levels in the brain and predicting the three-dimensional structure of the mutant protein. These analyses confirmed that Cyp19a1b function is indeed lost, thereby increasing the reliability of our conclusions.
FigS1 - panels C&D please indicate in which tissue were hormones measured. Blood?
We thank the reviewer for pointing this out. In our study, “peripheral” refers to the caudal half of the body excluding the head and visceral organs, not blood. Accordingly, we have revised the figure legend and the description in the Materials and Methods section as follows:
Legend for Figure 1B (line 787) now reads: “Levels of E2, testosterone, and 11KT in the brain (A) and peripheral tissues (caudal half of the body) (B) of adult cyp19a1b<sup>+/+</sup>, cyp19a1b<sup>+/−</sup>, and cyp19a1b<sup>−/−</sup> males (n = 3 per genotype).”
Materials and methods (line 431): The sentence “Total lipids were extracted from the brain and peripheral tissues (from the caudal half) of” has been revised to “Total lipids were extracted from the brain and from peripheral tissues, specifically the caudal half of the body excluding the head and visceral organs, of.”
Additional Alterations:
We have reformatted the text and supporting materials to comply with the journal’s Author Guidelines. The following changes have been made:
(1) Figures and supplementary files are now provided separately from the main text.
(2) The title page has been reformatted without any changes to its content.
(3) In-text citations have been changed from numerical references to the author–year format.
(4) Figure labels have been revised from “Fig. 1,” “Fig. S1,” etc., to “Figure 1,” “Figure 1—figure supplement 1,” etc.
(5) Table labels have been revised from “Table S1,” etc., to “Supplementary file 1,” etc.
(6) Line 324: The typo “is” has been corrected to “are”.
(7) Line 382: The section heading “Materials and Methods” has been changed to “Materials and methods” (lowercase “m”).
(8) Line 383: The Key Resources Table has been placed at the beginning of the Materials and methods section.
(9) Line 389: The sentence “Sexually mature adults (2–6 months) were used for experiments, and tissues were consistently sampled 1–5 hours after lights on.” has been revised to “Sexually mature adults (2–6 months) were used for experiments and assigned randomly to experimental groups. Tissues were consistently sampled 1–5 hours after lights on.”
(10) Line 393: The sentence “All fish were handled in accordance with the guidelines of the Institutional Animal Care and Use Committee of the University of Tokyo.” has been removed.
(11) Line 589: The following sentence has been added: “No power analysis was conducted due to the lack of relevant data; sample size was estimated based on previous studies reporting inter-individual variation in behavior and neural gene expression in medaka.”
(12) Line 598: The reference list has been reordered from numerical sequence to alphabetical order by author.
(13) In the figure legends, notations such as “A and B” have been revised to “A, B.”
-
-
www.gutenberg.org www.gutenberg.org
-
She didn’t like it,” he insisted. “She didn’t have a good time.” He was silent, and I guessed at his unutterable depression. “I feel far away from her,” he said. “It’s hard to make her understand.” “You mean about the dance?” “The dance?” He dismissed all the dances he had given with a snap of his fingers. “Old sport, the dance is unimportant.” He wanted nothing less of Daisy than that she should go to Tom and say: “I never loved you.” After she had obliterated four years with that sentence they could decide upon the more practical measures to be taken. One of them was that, after she was free, they were to go back to Louisville and be married from her house—just as if it were five years ago. “And she doesn’t understand,” he said. “She used to be able to understand. We’d sit for hours—” He broke off and began to walk up and down a desolate path of fruit rinds and discarded favours and crushed flowers.
yess even gastby sees daisy inside , i don;t get it why tom ignore her
-
-
Local file Local file
-
Some researchers look at someone creating analternative history in a game, like a small country conquering the world,and see it that way. But my argument is that these types of alternativehistory creation, or counter-play, is essentially just replicating the logicof colonialism. You just happen to be the colonizer. Like, let me go con-quer England in the game – it’s still colonialism, right?
The point being, there is exogenous, or what is forced from outside, and endogenous, or what is forced from inside. UK, France, Spain, etc. were and are colonialists countries that forced upon others their empire, and they still do, but in a more palatable less visibly violent fashion.
They opress, homogenise, silence, displace, much like the US, Mainland China, India, or Mexico. Their internal dissidents, varied ideologies, immigrants, subcultures, languages, natives, are squished, minoritised, colonised.
-
This perspective on technology as an unproblematic labor saving de-vice fits well with so-called common-sense but wrongheaded ideasabout technologies as neutral tools (see Myth #1) that can smoothlyand easily take on the burden of labor from humans and increase ef-ficiency. This idea has been notably critiqued by Langdon Winner butalso many other scholars of Science and Technology Studies such asBruno Latour (1996) and Susan Leigh Star (1999)
In a way, like with energy, which is not spent or generated but continuously transferred, we should not think in close yes-no, action-result. The event is part of a system, it's on the move, and efficiency doesn't emerge from nothing, it requires other work. I am not talking about zero-sum competition, we can most win with tech, but transformations like eye glasses or leg prosthetics need of workers on the other end, but by automatising them, we are just making them less visible, we are moving them from the artisan workshop to the factory or the mine. We'll have to wait a lot until this manual labour gets replaced by robotics, because once again, the trade-off is not "efficient" right now.
-
-
journals-scholarsportal-info.myaccess.library.utoronto.ca journals-scholarsportal-info.myaccess.library.utoronto.ca
-
Indeed, homes are spaces where the intimacy among family members can open up daily opportunities to “see, hear, think and act differently” (Armstrong, 1995) as we redefine what it means to decolonize and to queer our homes, bodies, and relationships.
Closing statement: home is where decolonial and queer group lives daily, it's important to see that decolonization happens within family settings rather than just political spaces.
-
K: I love you Mommy.C: I love you too.K: Do you love everyone in our family?C: Of course! I love you, Mum, Rosebud, Nana, all of our aunts, uncles, cousins, nephews, and our friends who are family to us, like Aunt Caroline.K: And our ancestors?C: Oh yes! I love our ancestors too.K: Well not all of them … we don't love our England ancestors who came over here and did the whole stealing the land from the Aboriginal people.C: Oh … those ancestors. Umm, right. Well, that's a really good question. Do we love people who do things that are wrong? Could we hate what they did, but love the people?K: Well, I’m not sure. I care about the Aboriginal people and what happened to them.C: I do too. That's a really interesting and important question you are asking.K: Yeah.C: It's not easy to figure that out, is it?K: No, it isn’t.
Decolonial work happens through kitchen tables, through parenting or just simple conversations.
-
Intimate relationships with partners or lovers raise a different set of questions about enacting decolonial practices across axes of race, gender, class, sexuality, and so on. As cisgender queer women, we have been in intimate relationships with genderqueer and trans partners in which we have enacted allyship by providing intimate support and care for partners encountering transphobia and heteronormativity in daily life. This requires undertaking self-education and self-reflection, particularly as our intimate relationships are spaces of reciprocal support in a different way than our friendships.
This part looks at romantic relationships as political spaces and shows that just like in family settings, how caregiving, labors are shaped by a much larger structure even if you think it's personal
-
We view “decolonization” and “queering” as active, interconnected, critical, and everyday practices that take place within and across diverse spaces and times.
It's more than just simple concepts; decolonization and queering are realities that people experience every day.
-
-
www.biorxiv.org www.biorxiv.org
-
Note: This response was posted by the corresponding author to Review Commons. The content has not been altered except for formatting.
Learn more at Review Commons
Reply to the reviewers
1. General Statements
In this study, we mechanistically define a new molecular interaction linking two of the cell's major morphological regulatory pathways-the Rho GTPase and Hippo signaling networks. These two major signaling pathways are both required for life across huge swaths of the tree of life. They are required for the dynamic organization and reorganization of proteins, lipids, and genetic material that occurs in essential cellular processes such as division, motility and differentiation. For decades these pathways have been almost exclusively studied independently, however, they are known to act in concert in cancer to drive cytoskeletal remodeling and morphological changes that promote proliferation and metastasis. However, mechanistic insight into how they are coordinated is lacking.
Our data reveal a mechanistic model where coordination is mediated by the RhoA GTPase-activating protein ARHGAP18, which forms molecular interactions with both the tumor suppressor Merlin (NF2) and the transcriptional co-regulator YAP (YAP1). Using a combination of state-of-the-art super-resolution microscopy (STORM, SORA-confocal) in cultured human cells, biochemical pulldown assays with purified proteins, and analyses of tissue-derived samples, we characterize ARHGAP18's function from the molecular to the tissue level in both native and cancer model systems.
Together, these findings establish a previously unrecognized molecular connection between the RhoA and Hippo pathways and culminate in a working model that integrates our current results with prior work from our group and decades of prior studies. This model provides a new conceptual framework for understanding how RhoA and Hippo signaling are coordinated to regulate cell morphology and tumor progression in human cells.
In this substantially revised manuscript, we have addressed all comments from the expert reviewers described point-by-point below. A shared major comment from the reviewers was the request for direct evidence of the proposed mechanistic model. To address these constructive comments, we've added new experiments, new quantification, new text, new control data, and have added two expert authors, adding super-resolution mouse tissue imaging data for the endogenous study of ARHGAP18 in its native condition. We believe that these additions greatly enhance the manuscript and collectively address the overall message from the reviewer's collective comments.
2. Point-by-point description of the revisions
Reviewer #1 (Evidence, reproducibility and clarity (Required)):
This manuscript describes a dual mechanism by which ARHGAP18 regulates the actin cytoskeleton. The authors propose that in addition to the known role for ARHGAP18 in regulating Rho GTPases, it also affects the cytoskeleton through regulation of the Hippo pathway transcriptional regulator YAP. ARHGAP18 knockout Jeg3 cells are were generated and show a clear loss of basal stress fiber like F-actin bundles. The authors further characterize the effects of ARHGAP18 knockout and overexpression. It is also discovered that ARHGAP18 binds to the Hippo pathway regulator Merlin and to YAP. Ultimately it is concluded that ARHGAP18 regulates the F-actin cytoskeleton through dual regulation of RHO GTPases and of YAP. While the phenotype of the ARHGAP18 knockout and the association of ARHGAP18 with Merlin and YAP is interesting, I found the authors conclusion that these phenotypes are due to ARHGAP18 regulation of both RHO and YAP to be based on largely correlative evidence and sometimes lacking in controls or tests for significance. In addition the authors often make overly strong conclusions based on the experimental evidence. In some instances, the rationale for how the experimental results support the conclusion is insufficiently articulated, making evaluation challenging. In general although the authors have some interesting observations, more definitive experiments with proper controls and statistical tests for significance and reproducibility are needed to justify their overall conclusions.
- *
*We appreciate the reviewers' constructive comments and have added substantial new data and quantifications to address their concerns. We have focused these new data on directly testing the proposed mechanisms, adding controls, and performing quantitative analysis with statistical testing. Additionally, we have edited our language to make our rationale clearer and to present our conclusions as a more moderate assessment of our experimental results. Below we respond to the specific comments made by the reviewer, followed by a list of additional editorial changes we've made based on the reviewer's overarching comments on clarity and rationale. *
Specific Comments
1) The authors make a big point about the effects of ARHGAP18 on myosin light chain phosphorylation. However, this result is not quantified and tested for statistical significance and reproducibility.
*We thank the reviewer for their comments on our western blotting quantification, which in the original submission version had quantification of RhoA downstream signaling of pCofilin/ Cofilin and pLIMK/ LIMK. We had withheld the pMLC and MLC quantification as the result was previously published with quantification, reproducibility, and statistical significance by our group in our prior manuscript on ARHGAP18 published in Elife in 2024 (Fig. 4E of *
https://doi.org/10.7554/eLife.83526 ). However, these prior results lacked the new overexpression data. We recognize the need to add these data to this manuscript as requested by the reviewer.
- *
*To address the reviewer's comment, we have added quantification of pMLC/MLC (Fig. 1F) *
2) Along similar lines in Figure 2C they state that overexpression of ARHGAP18 causes cells to invade over the top of their neighbors. This might be true and interesting, but only a single cell is shown and there is no quantification or controls for simply overexpressing something in that cell. The authors also conclude from this image that the overexpression phenotype is independent of its GAP activity on Rho. It is not clear how this conclusion is made based on the data. It would seem like a more definitive experiment would be to see if a similar phenotype was induced by an ARHGAP18 mutant deficient in GAP activity.
Based on the reviewer's comment, we recognize the qualitative statements made in Figure 2C (now Figure 3) should've been made more quantitative. We have added the control of Jeg 3 WT cells expressed with empty vector flag to show that WT cells do not invade over the top of each other (Fig. 3F). Additionally, we have added the quantification found in Fig. 3E, which shows the % invasive/ non-invasive cells between WT and ARHGAP18 overexpression cells. We have clarified our conclusions to make clear that these data do not directly test if the invasive phenotype derives from a Rho-independent mechanism. The text now states the following conclusion alongside others, which can be seen in our tracked changes:
- *
"These data support the conclusion that ARHGAP18 acts to regulate basal and junctional actin. However, it was not clear whether this activity occurred through a Rho-independent or a Rho-dependent mechanism."
- *
We have added new data of cells expressing an ARHGAP18 mutant deficient in GAP activity, which is explained in detail in the following response below.
3) In Figure 3 the authors compare gene expression profiles of ARHGAP18 knockout cells to wild-type cells. They see lots of differences in focal adhesion and cytoskeletal proteins and conclude that this supports their conclusion that ARHGAP18 is not just acting through RHO. The rationale for this in not clear. In addition, they observe changes in expression profiles consistent with changes in YAP activity. They conclude that the effects are direct. This very well might be true. However RHO is a potent regulator of YAP activity and the results seem quite consistent with ARHGAP18 acting through RHO to affect YAP.
- *
We thank the reviewer for their comment and believe the revised manuscript now presents direct evidence to support the conclusions made through the editing text and the incorporation of new data.
- *
First, the reviewer highlighted that we were not clear in our rationale and explanation of the conclusions made from our RNAseq data in the new Figure 4 (Previously Figure 3). We agree with the reviewer that the RNAseq data alone is not sufficient rationale for the conclusion that ARHGAP18 is acting through YAP directly. In the revised manuscript, the conclusion is now made based on the combination of our multi-faceted investigation of the relationship between ARHGAP18 and YAP (most importantly, new Figure 5). It's important for us to argue that our RNAseq analysis is much more robust and specific than simply reporting a descriptive assay seeing lots of differences in cytoskeletal proteins. We recruited an outside RNAseq expert collaborator; Dr. Yongho Bae, to perform state-of-the-art IPA analysis and a grueling manual curation of the top hit genes to identify the predominant signaling pathways linking the loss of ARHGAP18 to known YAP translational products. We've provided a supplemental table listing each citation supporting the identified YAP pathway associations from this manual curation. We also have added a new discussion paragraph on RNAseq data to clarify our specific RNAseq data results and analysis. In the revised manuscript, we have moderated our language in the results text regarding the RNAseq data to reflect the reviewer's suggestion:
- *
"Our RNAseq data alone could not independently confirm if the alterations to transcriptional signaling and expression of actin cytoskeleton proteins were through a Rho-dependent or Rho-independent mechanism."
-
*
-
*
Second, in this comment and the above, the reviewer highlights the need for a new experiment to directly test the Rho Independent effects of ARHGAP18, which we now provide in the new Figure 5. In this new data, we've applied an experimental design suggested by reviewer 2 regarding the same concern. In short, we've produced and expressed a point mutant variant ARHGAP18(R365A), which abolishes the Rho GAP activity while maintaining the remainder of the protein intact. This construct allows us to directly test the effects of ARHGAP18 independent from its RhoA GAP activity. We find that the GAP-deficient ARHGAP18 is able to fully rescue basal focal adhesions, indicating that the basal actin phenotype is at least in part regulated through a Rho-independent mechanism.
-
*
-
*
*We believe the revised manuscript, when taken in totality, provides the definitive proof requested by the reviewer. Specifically, the combination of Figure 5, where we show new data using the ARHGAP18(R365A) variant, and the result that ARHGAP18 forms a stable complex with YAP (Fig. 6G) or Merlin (Fig.6A), is supportive of direct Rho-independent molecular interactions between YAP, Merlin, and ARHGAP18. *
4) In Figure 4A showing Merlin binding to ARHGAP18 there is no control for the amount of Merlin sticking to the column as was done in Figure 4F for binding experiments with YAP. This makes it difficult to determine the significance of the observed binding.
We have performed the requested control experiment and added the results to Figure 6A.
5) The images in Figure 4C showing YAP being maintained in the nucleus more in ARHGAP18 knockout cells compared to wild-type. However the images only show a few cells and YAP localization can be highly variable depending on where you look in a field. Images with more cells and some sort of quantification would bolster this result.
We have provided quantification (Figure 6D) of what was originally Figure 4C (now Figure 6C).
Reviewer #1 (Significance (Required)):
While the phenotype of the ARHGAP18 knockout and the association of ARHGAP18 with Merlin and YAP is interesting, I found the authors conclusion that these phenotypes are due to ARHGAP18 regulation of both RHO and YAP to be based on largely correlative evidence and sometimes lacking in controls or tests for significance. In addition the authors often make overly strong conclusions based on the experimental evidence. In some instances, the rationale for how the experimental results support the conclusion is insufficiently articulated, making evaluation challenging. In general although the authors have some interesting observations, more definitive experiments with proper controls and statistical tests for significance and reproducibility are needed to justify their overall conclusions.
In the above comments, we detail the specific definitive experiments, proper controls, and statistical tests for significance, requested by the reviewer, which we believe greatly strengthen our manuscript.
Reviewer #2 (Evidence, reproducibility and clarity (Required)):
This manuscript investigates the Rho effector, ARHGAP18 in Jegs cells, a trophoblastic cell line. It presents a number of new pieces of data, which increase our understanding of the importance of this GAP on cell function and explains at a molecular level previous results of other workers in the field. ARHGAP18 was originally given the name "conundrum' and continues to stand apart from the majority of other GAP proteins and their functions. Hence the data here is significant and of high standard.
The data is clear, and the images are of high quality and extremely impressive in their resolution. It is significant and adds a further layer to our understanding of the regulation of cell migration, particularly in the formation and resolution of microvilli.
- *
We appreciate the reviewer's comments and supportive insights.
The data is based on the use of the cell line Jeg3. Even the authors previous publication in eLife is based only on this cell line. They need to show the conclusions are general and not specific to this line of cells. As an extension of this, is the ARHGAP18 function shown here only in transformed cells? Does the same mechanisms operate in normal cells, which respond to activation to proliferate or migrate?
- *
- We respectfully point out that the critical experiments of the prior eLife publication were validated in DLD-1 colorectal cells and not Jeg-3 cells alone (Figure 1-figure supplement 2). Our newly independent lab, established just over a year ago, is unable to perform a full expansion of the manuscript using untransformed cells, however, we agree with the reviewer's perspective and wish to address the comment to the best of our current capability. To answer the reviewers' suggestions, we have recruited Dr. Christine Schaner Tooley, an expert in mouse model system studies. In the revised manuscript, we've added new Super-Resolution SORA confocal images of endogenous ARHGAP18's localization in the intact intestinal villi tissue, and apical junctions of WT mice (Fig.1A-C). These data indicate that endogenous ARHGAP18 is enriched (but not exclusively localized) at the apical plasma membranes of normal WT epithelial cells. This localization, where both Merlin and Ezrin are present at apical membrane/ junctions under normal conditions, is a major component of the working model proposed in Fig. 7. These data also indicate that ARHGAP18 is capable of entering the nucleus in WT cells, another critical aspect of our proposed model. Collectively, our DLD-1 studies published previously and or new studies using WT mice tissue samples support the conclusion that at least some of ARHGAP18's functions described in this manuscript are not limited to Jeg3 cells.*
In endothelial cells, Lovelace et al 2017 showed localization to microtubules and that depletion of ARHGAP18 resulted in microtubule instability. The authors may like to comment on the differences. Is this a cell type difference or RhoA versus RhoC difference?
- *
In our previous publication (Lombardo Elife), we validated the finding that ARHGAP18 forms a complex with microtubules, as we detected tubulin in the ARHGAP18 pulldown experiment (Figure 1- Source Data). However, our data indicate that in Jeg3 cells ARHGAP18 does not localize to the same microtubule associated spheres observed in the Lovelace publication. We now comment on the shared conclusions and differences between this manuscript and the Lovelace et al 2017 in the discussion section.
- *
"In endothelial cells, ARHGAP18 has been reported to localize microtubules and plays a role in maintaining proper microtubule stability (Lovelace et al., 2017). In our epithelial cell culture models and WT mouse intestine, we have been unable to detect ARHGAP18 at microtubules suggesting ARHGAP18 may have additional functions is various cell types."
On pages 7,9 they conclude that MLC and basal and junctional actin are regulated through a GAP independent mechanism. The best way to show this is with overexpression of a GAP mutant.
We appreciate the reviewer's insight and have produced and expressed a GAP mutant, ARHGAP18(R365A), in our cells, directly testing our conclusion that ARHGAP18 has a GAP-independent function. These data are now presented in revised Figure 5 and explained further in response to reviewer #1.
There is a huge amount of data presented in Figure 3, but their 2 genes which they focus on, LOP1 and CORO1A, are discussed but no actual data presented in support.
We now validate the CORO1A by qPCR in Figure 4J.
- *
Reviewer #2 (Significance (Required)):
The data is significant and adds a further layer to our understanding of the regulation of cell migration, particularly in the formation and resolution of microvilli. This manuscript will be of significance to an basic science audience in the field of RhoGTPases and cell migration.
Reviewer #3 (Evidence, reproducibility and clarity (Required)):
The study by Murray et al explores the effects of ARHGAP18 on the actin cytoskeleton, Rho effector kinases, non-muscle myosin, and transcription. Using super resolution microscopy, they show that in ARHGAP18 KO cells there is a mixed and unexpected cytoskeleton phenotype where myosin phosphorylation appears to be increased, but actin is disorganised with reduced stress fibres, diminished focal adhesions and augmented invasiveness. They conclude that the underlying mechanisms are likely independent from RhoA. Next, they perform RNAseq using the KO cells and identify an array of dysregulated genes, including those that play crucial roles in microvilli (related to previously published findings). Analysis of the data identify gene expression changes that are relevant for altered focal adhesion (integrins). Further analysis reveals that a large cohort of the dysregulated genes are YAP targets. They then show that in ARHGAP18 KO cells YAP nuclear localization, as detected by immunostaining, is augmented; and demonstrate that immobilized ARHGAP18 protein can bind the Hippo regulator merlin as well as YAP itself.
Major comments:
1, The premise of the study (that ARHGAP18 is a RhoA effector or may acts independently of RhoA) remains not proven.
We have added new evidence of direct RhoA independent activity for ARHGAP18 described in the above comments. Specifically, we've added data using a RhoA-GAP dead variant of ARHGAP18 in Figure 5, which we believe addresses this comment.
- *
At several places (including in the title) the authors refer to ARHGAP18 as a Rho effector, which would suggest that it is downstream form Rho, but the basis for this is not clear. In fact, their own previous study suggested that ARHGAP is a RhoA regulator, rather than an effector. In general, the connection of the described effects to RhoA remains unclear, and not addressed in this study. The authors seem to go back and forth in their conclusions regarding the connection between ARHGAP18 and RhoA. For example, the first section of results is finished by stating (line 194): "These data support the conclusion that ARHGAP18 acts to regulate basal and junctional actin through Rho-independent mechanism". But the next section starts by stating (line 198): "We hypothesized that the invasive and cytoskeletal phenotypes observed at the basal surface of cells devoid of ARHGAP18 may be a result of changes in regulation at the transcriptional level either directly through RhoA signaling or through an additional mechanism specific to ARHGAP18". The paper would be strengthened by adding data that show whether the effects are indeed downstream, from RhoA or RhoA independent. If there is no sufficient demonstration that ARHGAP18 is downstream of RhoA and is an effector, this needs to be stated explicitly, and the wording should be changed.
*We now provide new data in Figure 5, which directly tests the RhoA independent functions of ARHGAP18 as recommended by the reviewer. Our understanding of the term effector is 'a molecule that activates, controls, or inactivates a process or action.' Based on this understanding, we used the term to convey ARHGAP18's functional role within the feedback loop, rather than to imply that it acts exclusively downstream. *
- *
We seek to clarify our perspective with the reviewer's assertion that we go "back and forth" as to if ARHGAP18 functions in a Rho Dependent or Rho Independent manner. It was our intent to propose a model where ARHGAP 18 acts in two separate circuits that regulate cell signaling. The first circuit involves ARHGAP18's canonical RhoA GAP activity, which involves ERMs and LOK/SLK, and is limited to the apical plasma membrane. This first signaling circuit was characterized in our prior Elife manuscript (Lombardo et al., 2024) and in an earlier JCB manuscript (Zaman and Lombardo et al., 2021). In this newly revised manuscript, we provide a partial mechanistic characterization of the second circuit, which we freely admit is much more complex and will likely require additional study to fully characterize.
- *
As both circuits operate as signaling feedback loops, we find the terms 'upstream' and 'downstream' to be of limited value, and we attempt to avoid their use when possible. We retain their use only when referring to the Hippo and ROCK signaling cascades, where these designations are well established. We suggest that the conceptual inconsistencies of Conundrum/ARHGAP18 may have arisen from the tendency to view it in strictly binary terms as upstream or downstream. Here, we propose a third possibility that ARHGAP18 functions as both, participating in a negative feedback loop.
- *
*We have edited and added data testing if the effects are Rho independent and discussion text in response to the reviewer's comments and clarify the molecular function of ARHGAP18.
"Additionally, focal adhesions and basal actin bundles are restored to WT levels when the ARHGAP18(R365A) GAP-ablated mutant is expressed in ARHGAP18 KO cells (Fig. 5A, B). These results represent the strongest argument that ARHGAP18 functions in additional pathways to RhoA/C alone. Our data suggests that at least one of the alternative pathways is through ARHGAP18's interaction with YAP and Merlin. From these data we conclude that ARHGAP18 has important functions in both RhoA signaling through both its GAP activity and in Hippo signaling through its GAP independent binding partners. "*
-
*
-
*
The study is descriptive and contains a series of observations that are not connected. Because of this, the study's conclusions are not well supported, and key mechanistic insight is limited. The study feels like a set of separate observations, that remain incompletely worked out and have some preliminary feel to them. The model in the last figure also seems to contain hypotheses based on the observations, several of which remains to be proven.
- *
*We present our revised manuscript, in which we've more clearly outlined our rationale and conclusions, as detailed in the above responses, to emphasize the overall connectivity of the study. We have also updated the title of Figure 7 to read "__Theoretical __Model of ARHGAP18's coordination of RhoA and Hippo signaling pathways in Human epithelial cells." To make it clear that we are presenting a working model, which has elements that will require additional investigation. Throughout the manuscript, we highlight the unknown elements that remain to be tested or other outstanding questions. Thus, we do not aim to characterize this complex signaling coordination completely. Instead, this manuscript represents the 3rd iteration in our systematic advances to describe this entirely new signaling pathway. We agree that, despite three separate manuscripts (this one included) to date, this work represents an early stage in understanding the system, many additional studies will be needed to characterize this signaling system fully. Figure 7 is presented as a working model that results from a thoughtful combination of our collective data and that of other researchers, derived from numerous species across decades of study. We firmly believe that proposing such integrative models is valuable for advancing the field. We also recognize the importance of clearly indicating which aspects remain hypothetical. We now explicitly note in several places within the discussion which components of the model will require further validation and experimental confirmation. For example, regarding our theoretical mechanism in Figure 7 we state: *
"Validation of the direct mechanism by which YAP/TAZ transcriptional changes drive basal actin changes in ARHGAP18 KO cells will require further investigation based on predictions from RNAseq results."
- *
Addressing any possible connection between key effects of ARHGAP18 KO (changes in actin, focal adhesion, integrins, Yap and merlin binding) could strengthen the manuscript. One such specific question is the whether the changes in integrin expression (RNAseq) are indeed connected to the actin alterations and reduction ion focal adhesions (Fig 1). Staining for these integrins to show they are indeed altered, and/or manipulating any of them to reproduce changes could provide and exciting addition.
- *
*We attempted to stain cells for Integrins by purchasing three separate antibodies. However, despite extensive optimization and careful selection of the specific integrins using our RNAseq results we were unable to get any of these antibodies to work in any cell type or condition. We believe that there is a technical challenge to staining for integrins due to their transmembrane and extracellular components, which we were unable to overcome. As an attempt to address the reviewers comment, we alternatively stained cells for paxillin which directly binds the cytoplasmic tails of integrins (Fig. 3&5). *
Some of the experimental findings are not convincing or lack controls. Fig 1: some of the western blots are not convincing or poor quality. [...] On the same figure, the quality of LIM kinase blots is poor. [...] The signal is weak, and the blot does not appear to support the quantification. The last condition (expression of flag-ARHGAP18) results in a large drop in pLIMK and pcofilin on the blot, which is not reflected by the graph. Addition of *a better blot and the use of strong positive or negative control would boost confidence in these data. *
- *
In response to this and other reviewers' comments, we have added new western data and quantification to Figure 1. We now focus on MLC/pMLC data as we believe these data highlight the potential Rho-independent mechanism of ARHGAP18, and we were able to greatly improve the quality of the blots through careful optimization. We hope the reviewer finds these blots and quantifications (Fig. 1E and F) more convincing.
*We note that phospho-specific Western blotting presents considerably greater technical challenges than conventional blotting. We believe that the appearance of an attractive looking blot does not always correlate to quality or reproducibility and have focused on taking extraordinarily careful steps in the blotting of our phospho-specific antibodies, which at times comes at the cost of the blot's attractiveness in appearance. For example, all phospho-specific antibodies are run using two color fluorescent markers to blot against both the total protein and the phospho-protein on the same blot. This approach often leads to blots that have reduced signal to noise compared to chemiluminescent Westerns. Additionally, we use phospho-specific blocking buffer reagents which do not contain phosphate-based buffers or agents that attract non-specific phospho-staining signals. These blocking buffers are not as effective as non-fat milk in pbs at blocking the background signal, however, they are ultimately cleaner for phospho-specific primary antibodies. We use carefully optimized protocols, from cell treatment to lysis, transfer, and antibody incubation, including methods developed by laboratories where the corresponding author of the manuscript was trained. Nonetheless, despite these efforts, we have now removed the LIMK and cofilin data because we deemed them unnecessary for the main conclusions of this manuscript and were unable to improve their quality to satisfy the reviewer. *
The changes in pMLC on the western blots are very small, and for any conclusion, these studies require quantification. Further, the expression levels of Flag-ARHGAP18 needs to be shown to support the statement that the protein is expressed, and indeed overexpressed under these conditions (vs just re-expressed).
In continuation of the above comment, we have made significant effort to improve the quality of our pMLC western blots and now provide quantification in Figure 1. We also now provide the Flag-ARHGAP18 signal as requested by the reviewer.
Fig 4: the differences in YAP nuclear localization under the various conditions are not well visible. Quantitation of nuclear/cytosolic signal ratio should be provided. Please provide a rationale and more context for using serum starvation and re-addition. What is the expected effect? Serum removal and addition is referred to as nutrient removal and re-addition, but this is inaccurate, as it does not equal nutrient removal, since serum contains a variety of other important components, e.g. growth factors too.
We have provided new quantification of the nuclear/cytosolic signal ratio in Figure 6D. We have explained our rational for the study through the following new text:
"Merlin is activated and localized to junctions upon signaling, promoting growth and proliferation; among these signals is the availability of growth factors and other components of serum (Bretscher et al., 2002). We hypothesized that since ARHGAP18 formed a complex with Merlin that ARHGAP18's localization may localize to junctions under conditions which promote Merlin activation."
- *
We have altered our use of "nutrient removal" to "serum removal"
The binding between ARHGAP18 and merlin is interesting, but a key limitation is the use of expressed proteins. Can the binding be shown for the endogenous proteins (IP, colocalization). Another important unaddressed question is the relevance of this binding, and the relation of this to altered YAP nuclear localization.
- *
*Our data in Fig. 6G shows binding of a resin bound human ARHGAP18 to endogenous YAP from human cells as suggested by the reviewer. In Fig. 6A, we have selected to use GFP-Merlin as Merlin shares approximately 60% sequence identity with Ezrin, Radixin, and Moesin (ERMs). Their similarity is such that Merlin was named for Moesin-Ezrin-Radixin-Like Protein. In our experience, nearly all Merlin or ERM antibodies have some cross-contaminating signal. Thus, a major concern is that if we were to blot for endogenous Merlin in the pull-down experiment, we may see a band that could in fact be ERMs. To avoid this, we tagged Merlin with GFP to ensure that the product pulled down by ARHGAP18 was Merlin, not an ERM. Regarding the ARHGAP18-resin bound column, our homemade ARHGAP18 antibody is polyclonal. We have extensive experience in pulldown assays and have found that the binding of a polyclonal antibody to the bait protein can produce less accurate results, as the binding site for the antibody is unknown and can sterically hinder attachment of target proteins like Merlin. In our experience, attachment to a flag-tag, which is expressed after a flexible linker at the N- or C-terminus, allows us to overcome this limitation, which we've used in this manuscript. *
Minor comments:
Introduction line 99: "When localized to the nucleus, YAP/TAZ promotes the activation of cytoskeletal transcription factors associated with cell proliferation and actin polymerization" Please clarify what you mean by this statement, that is inaccurate in its present for. Did you mean effects on transcription factors that control cytoskeletal proteins, or do you mean that Yap/Taz affect these proteins? Please also provide reference for this.
We've altered the sentence as suggested by the reviewer, which now reads the following:
"When localized to the nucleus, YAP/TAZ promotes transcriptional changes associated with cell proliferation and actin polymerization."
- *
*The full mechanism for how YAP/TAZ promotes proliferation and actin polymerization is a currently debated issue. We do not think introducing the various current proposed models is required for this manuscript, and we simply intend to convey that when in the nucleus, YAP/TAZ promotes transcriptional changes that drive actin polymerization and cell proliferation. *
-What is the cell confluence in these experiments? For epithelial cells confluence affects actin structure. Please comment on similarity of confluency across experimental conditions?
- *
All cellular experiments are paired where WT and ARHGAP18 KO cells are plated at the same time under identical conditions. For imaging, we plate all cells onto glass coverslips in a 6 well dish so that each condition is literally in the same cell culture plate and gets identical treatment. In our prior Elife paper studying ARHGAP18, we characterized that ARHGAP18 KO cells and WT cells divide at a similar rate and have similar proliferation characteristics. The epithelial cell cultures are maintained for experiments around 70-80% confluency. For the focal adhesion staining experiments, the confluency is slightly lower, between 50-60% to capture the focal adhesions towards the leading edge. We have added the following new text to further describe these methods: "Cell cultures for experiments were maintained at 70%-80% confluency. For focal adhesion experiments, the cell cultures were maintained at 50%-60% confluency."
-Fig 2 legend: please indicate that the protein detected was non-muscle myosin heavy chain (distinct from the light chain detected in Fig 1).
- *
We have altered original Figure 2 (new Figure 3) legend.
-Line 339-340: please check the syntax of this sentence -Western blot quantification: the comparison of experiments with samples run on different gels/blots requires careful normalization and experimental consistency. Please describe how this was achieved.
- *
We have added the following new text to further describe these methods:
"For blots which required quantification of antibodies that were only rabbit primaries (e.g., pMLC/MLC antibodies listed above), samples were loaded onto a single gel and transferred onto a single membrane at the same time. After transfer, the membrane was cut in half and subsequent steps were done in parallel. All quantified blots were checked for equal loading using either anti-tubulin as a housekeeping protein or total protein as detected by Coomassie staining"
Reviewer #3 (Significance (Required)):
Rho signalling is a central regulator of an array of normal and pathological cell functions, and our understanding of the context dependent regulation of this key pathway remains very incomplete. Therefore, new knowledge on the role of specific regulators, such as ARHGAP18, is of interest to a very broad range of researchers. A further exciting aspect of this protein, that despite indications by many studies that it acts as a GAP (inhibitor) for Rho proteins, there are findings in the literature that suggest that its manipulation can affect actin in unexpected (opposite) manner. These point to possible Rho-independent roles, and warranted further in-depth exploration.
One of the strength of the study is that it explores possible roles of ARHGAP18 beyond RhoA and describes some new and interesting observations, which advance our knowledge. The authors use some excellent tools (e.g. ARHGAP KO cells and re-expression) and approaches (e.g. super resolution microscopy to analyze actin changes, RNAseq and bioinformatics to find genes that may be downstream from ARHGAP18). A key limitation of the study however, is that it is not clear whether the observed findings are indeed independent from RhoA. Further limitation is that potential causal relationships between the described findings are not studied, and therefore the findings are in some cases overinterpreted, and limited mechanistic insights are provided. In some cases the exclusive use of expressed proteins is also a limitation. Finally, some of the experiments also need improvement.
Reviewer expertise: RhoA signalling, guanine nucleotide exchange factors, epithelial biology, cell migration, intercellular junctions.
In the above comments, we detail the new experimental data addressing reviewer 3's listed key limitations. We've added new data using the Rho GAP deficient ARHGAP18(R365A) variant which allows for the direct characterization of ARHGAP18's Rho independent activity. We have introduced new data in WT cells studying endogenous proteins to address the limitations from expressed proteins. Finally, we have moderated our language to address overinterpretation. Collectively, we believe that our revised manuscript addresses the constructive reviewer's comments.
-
-
sites.google.com sites.google.com
-
“I’ve just always had the idea of growing and continuing to expand and show who I am outside of the basketball court,”
I find this so fascinating. I feel like entrepreneurs are always just extremely creative or athletic. For example, the owner of Adidas was a huge sports fan that he even made a shoe brand for athletes. It's cool to see how his love for a sport translated into something bigger like owning a business. I had no idea an ice cream company had so much depth to it.
-
-
www.biorxiv.org www.biorxiv.org
-
Reviewer #1 (Public review):
Summary:
In this manuscript, the authors investigate the nanoscopic distribution of glycine receptor subunits in the hippocampus, dorsal striatum, and ventral striatum of the mouse brain using single-molecule localization microscopy (SMLM). They demonstrate that only a small number of glycine receptors are localized at hippocampal inhibitory synapses. Using dual-color SMLM, they further show that clusters of glycine receptors are predominantly localized within gephyrin-positive synapses. A comparison between the dorsal and ventral striatum reveals that the ventral striatum contains approximately eight times more glycine receptors and this finding is consistent with electrophysiological data on postsynaptic inhibitory currents. Finally, using cultured hippocampal neurons, they examine the differential synaptic localization of glycine receptor subunits (α1, α2, and β). This study is significant as it provides insights into the nanoscopic localization patterns of glycine receptors in brain regions where this protein is expressed at low levels. Additionally, the study demonstrates the different localization patterns of GlyR in distinct striatal regions and its physiological relevance using SMLM and electrophysiological experiments. However, several concerns should be addressed.
Specific comments on the original version:
(1) Colocalization analysis in Figure 1A. The colocalization between Sylite and mEos-GlyRβ appears to be quite low. It is essential to assess whether the observed colocalization is not due to random overlap. The authors should consider quantifying colocalization using statistical methods, such as a pixel shift analysis, to determine whether colocalization frequencies remain similar after artificially displacing one of the channels.
(2) Inconsistency between Figure 3A and 3B. While Figure 3B indicates an ~8-fold difference in the number of mEos4b-GlyRβ detections per synapse between the dorsal and ventral striatum, Figure 3A does not appear to show a pronounced difference in the localization of mEos4b-GlyRβ on Sylite puncta between these two regions. If the images presented in Figure 3A are not representative, the authors should consider replacing them with more representative examples or providing an expanded images with multiple representative examples. Alternatively, if this inconsistency can be explained by differences in spot density within clusters, the authors should explain that.
(3) Quantification in Figure 5. It is recommended that the authors provide quantitative data on cluster formation and colocalization with Sylite puncta in Figure 5 to support their qualitative observations.
(4) Potential for pseudo replication. It's not clear whether they're performing stats tests across biological replica, images, or even synapses. They often quote mean +/- SEM with n = 1000s, and so does that mean they're doing tests on those 1000s? Need to clarify.
(5) Does mEoS effect expression levels or function of the protein? Can't see any experiments done to confirm this. Could suggest WB on homogenate, or mass spec?
(6) Quantification of protein numbers is challenging with SMLM. Issues include i) some of FP not correctly folded/mature, and ii) dependence of localisation rate on instrument, excitation/illumination intensities, and also the thresholds used in analysis. Can the authors compare with another protein that has known expression levels- e.g. PSD95? This is quite an ask, but if they could show copy number of something known to compare with, it would be useful.
(7) Rationale for doing nanobody dSTORM not clear at all. They don't explain the reason for doing the dSTORM experiments. Why not just rely on PALM for coincidence measurements, rather than tagging mEoS with a nanobody, and then doing dSTORM with that? Can they explain? Is it to get extra localisations- i.e. multiple per nanobody? If so, localising same FP multiple times wouldn't improve resolution. Also, no controls for nanobody dSTORM experiments- what about non-spec nb, or use on WT sections?
(8) What resolutions/precisions were obtained in SMLM experiments? Should perform Fourier Ring Correlation (FRC) on SR images to state resolutions obtained (particularly useful for when they're presenting distance histograms, as this will be dependent on resolution). Likewise for precision, what was mean precision? Can they show histograms of localisation precision.
(9) Why were DBSCAN parameters selected? How can they rule out multiple localisations per fluor? If low copy numbers (<10), then why bother with DBSCAN? Could just measure distance to each one.
(10) For microscopy experiment methods, state power densities, not % or "nominal power".
(11) In general, not much data presented. Any SI file with extra images etc.?
(12) Clarification of the discussion on GlyR expression and synaptic localization: The discussion on GlyR expression, complex formation, and synaptic localization is sometimes unclear, and needs terminological distinctions between "expression level", "complex formation" and "synaptic localization". For example, the authors state: "What then is the reason for the low protein expression of GlyRβ? One possibility is that the assembly of mature heteropentameric GlyR complexes depends critically on the expression of endogenous GlyR α subunits." Does this mean that GlyRβ proteins that fail to form complexes with GlyRα subunits are unstable and subject to rapid degradation? If so, the authors should clarify this point. The statement "This raises the interesting possibility that synaptic GlyRs may depend specifically on the concomitant expression of both α1 and β transcripts." suggests a dependency on α1 and β transcripts. However, is the authors' focus on synaptic localization or overall protein expression levels? If this means synaptic localization, it would be beneficial to state this explicitly to avoid confusion. To improve clarity, the authors should carefully distinguish between these different aspects of GlyR biology throughout the discussion. Additionally, a schematic diagram illustrating these processes would be highly beneficial for readers.
(13) Interpretation of GlyR localization in the context of nanodomains. The distribution of GlyR molecules on inhibitory synapses appears to be non-homogeneous, instead forming nanoclusters or nanodomains, similar to many other synaptic proteins. It is important to interpret GlyR localization in the context of nanodomain organization.
Significance:
The paper presents biological and technical advances. The biological insights revolve mostly on the documentation of Glycine receptors in particular synapses in forebrain, where they are typically expressed at very low levels. The authors provide compelling data indicating that the expression is of physiological significance. The authors have done a nice job of combining genetically tagged mice with advanced microscopy methods to tackle the question of distributions of synaptic proteins. Overall, these advances are more incremental than groundbreaking.
Comments on revised version:
The authors have addressed the majority of the significant issues raised in the review and revised the manuscript appropriately. One issue that can be further addressed relates to the issue of pseudo-replication. The authors state in their response that "All experiments were repeated at least twice to ensure reproducibility (N independent experiments). Statistical tests were performed on pooled data across the biological replicates; n denotes the number of data points used for testing (e.g., number of synaptic clusters, detections, cells, as specified in each case).". This suggests that they're not doing their stats on biological replicates, and instead are pseudo replicating. It's not clear how they have ensured reproducibility, when the stats seem to have been done on pooled data across repeats.
-
Author response:
The following is the authors’ response to the current reviews.
We thank the editors of eLife and the reviewers for their thorough evaluation of our study. As regards the final comments of reviewer 1 please note that all experimental replicates were first analyzed separately, and were then pooled, since the observed changes were comparable between experiments. This mean that statistical analyses were done on pooled biological replicates.
The following is the authors’ response to the original reviews.
General Statements
We thank the reviewers for their thorough and constructive evaluation of our work. We have revised the manuscript carefully and addressed all the criticisms raised, in particular the issues mentioned by several of the reviewers (see point-by-point response below). We have also added a number of explanations in the text for the sake of clarity, while trying to keep the manuscript as concise as possible.
In our view, the novelty of our research is two-fold. From a neurobiological point of view, we provide conclusive evidence for the existence of glycine receptors (GlyRs) at inhibitory synapses in various brain regions including the hippocampus, dentate gyrus and sub-regions of the striatum. This solves several open questions and has fundamental implications for our understanding of the organisation and function of inhibitory synapses in the telencephalon. Secondly, our study makes use of the unique sensitivity of single molecule localisation microscopy (SMLM) to identify low protein copy numbers. This is a new way to think about SMLM as it goes beyond a mere structural characterisation and towards a quantitative assessment of synaptic protein assemblies.
Point-by-point description of the revisions
Reviewer #1 (Evidence, reproducibility and clarity):
In this manuscript, the authors investigate the nanoscopic distribution of glycine receptor subunits in the hippocampus, dorsal striatum, and ventral striatum of the mouse brain using single-molecule localization microscopy (SMLM). They demonstrate that only a small number of glycine receptors are localized at hippocampal inhibitory synapses. Using dual-color SMLM, they further show that clusters of glycine receptors are predominantly localized within gephyrinpositive synapses. A comparison between the dorsal and ventral striatum reveals that the ventral striatum contains approximately eight times more glycine receptors and this finding is consistent with electrophysiological data on postsynaptic inhibitory currents. Finally, using cultured hippocampal neurons, they examine the differential synaptic localization of glycine receptor subunits (α1, α2, and β). This study is significant as it provides insights into the nanoscopic localization patterns of glycine receptors in brain regions where this protein is expressed at low levels. Additionally, the study demonstrates the different localization patterns of GlyR in distinct striatal regions and its physiological relevance using SMLM and electrophysiological experiments. However, several concerns should be addressed.
The following are specific comments:
(1) Colocalization analysis in Figure 1A. The colocalization between Sylite and mEos-GlyRβ appears to be quite low. It is essential to assess whether the observed colocalization is not due to random overlap. The authors should consider quantifying colocalization using statistical methods, such as a pixel shift analysis, to determine whether colocalization frequencies remain similar after artificially displacing one of the channels.
Following the suggestion of reviewer 1, we re-analysed CA3 images of Glrb<sup>eos/eos</sup> hippocampal slices by applying a pixel-shift type of control, in which the Sylite channel (in far red) was horizontally flipped relative to the mEos4b-GlyRβ channel (in green, see Methods). As expected, the number of mEos4b-GlyRβ detections per gephyrin cluster was markedly reduced compared to the original analysis (revised Fig. 1B), confirming that the synaptic mEos4b detections exceed chance levels (see page 5).
(2) Inconsistency between Figure 3A and 3B. While Figure 3B indicates an ~8-fold difference in the number of mEos4b-GlyRβ detections per synapse between the dorsal and ventral striatum, Figure 3A does not appear to show a pronounced difference in the localization of mEos4bGlyRβ on Sylite puncta between these two regions. If the images presented in Figure 3A are not representative, the authors should consider replacing them with more representative examples or providing an expanded images with multiple representative examples. Alternatively, if this inconsistency can be explained by differences in spot density within clusters, the authors should explain that.
The pointillist images in Fig. 3A are essentially binary (red-black). Therefore, the density of detections at synapses cannot be easily judged by eye. For clarity, the original images in Fig. 3A have been replaced with two other examples that better reflect the different detection numbers in the dorsal and ventral striatum.
(3) Quantification in Figure 5. It is recommended that the authors provide quantitative data on cluster formation and colocalization with Sylite puncta in Figure 5 to support their qualitative observations.
This is an important point that was also raised by the other reviewers. We have performed additional experiments to increase the data volume for analysis. For quantification, we used two approaches. First, we counted the percentage of infected cells in which synaptic localisation of the recombinant receptor subunit was observed (Fig. 5C). We found that mEos4b-GlyRa1 consistently localises at synapses, indicating that all cells express endogenous GlyRb. When neurons were infected with mEos4b-GlyRb, fewer cells had synaptic clusters, meaning that indeed, GlyR alpha subunits are the limiting factor for synaptic targeting. In cultures infected with mEos4b-GlyRa2, only very few neurons displayed synaptic localisation (as judged by epifluorescence imaging). We think this shows that GlyRa2 is less capable of forming heteromeric complexes than GlyRa1, in line with our previous interpretation (see pp. 9-10, 13).
Secondly, we quantified the total intensity of each subunit at gephyrin-positive domains, both in infected neurons as well as non-infected control cultures (Fig. 5D). We observed that mEos4bGlyRa1 intensity at gephyrin puncta was higher than that of the other subunits, again pointing to efficient synaptic targeting of GlyRa1. Gephyrin cluster intensities (Sylite labelling) were not significantly different in GlyRb and GlyRa2 expressing neurons compared to the uninfected control, indicating that the lentiviral expression of recombinant subunits does not fundamentally alter the size of mixed inhibitory synapses in hippocampal neurons. Interestingly, gephyrin levels were slightly higher in hippocampal neurons expressing mEos4b-GlyRa1. In our view, this comes from an enhanced expression and synaptic targeting of mEos4b-GlyRa1 heteromers with endogenous GlyRb, pointing to a structural role of GlyRa1/b in hippocampal synapses (pp. 10, 13).
The new data and analyses have been described and illustrated in the relevant sections of the manuscript.
(4) Potential for pseudo replication. It's not clear whether they're performing stats tests across biological replica, images, or even synapses. They often quote mean +/- SEM with n = 1000s, and so does that mean they're doing tests on those 1000s? Need to clarify.
All experiments were repeated at least twice to ensure reproducibility (N independent experiments). Statistical tests were performed on pooled data across the biological replicates; n denotes the number of data points used for testing (e.g., number of synaptic clusters, detections, cells, as specified in each case). We have systematically given these numbers in the revised manuscript (n, N, and other experimental parameters such as the number of animals used, coverslips, images or cells). Data are generally given as mean +/- SEM or as mean +/- SD as indicated.
(5) Does mEoS effect expression levels or function of the protein? Can't see any experiments done to confirm this. Could suggest WB on homogenate, or mass spec?
The Glrb<sup>eos/eos</sup> knock-in mouse line has been characterised previously and does not to display any ultrastructural or functional deficits at inhibitory synapses (Maynard et al. 2021 eLife). GlyRβ expression and glycine-evoked responses were not significantly different to those of the wildtype. The synaptic localisation of mEos4b-GlyRb in KI animals demonstrates correct assembly of heteromeric GlyRs and synaptic targeting. Accordingly, the animals do not display any obvious phenotype. We have clarified this in the manuscript (p. 4). In the case of cultured neurons, long-term expression of fluorescent receptor subunits with lentivirus has proven ideal to achieve efficient synaptic targeting. The low and continuous supply of recombinant receptors ensures assembly with endogenous subunits to form heteropentameric receptor complexes (e.g. [Patrizio et al. 2017 Sci Rep]). In the present study, lentivirus infection did not induce any obvious differences in the number or size of inhibitory synapses compared to control neurons, as judged by Sylite labelling of synaptic gephyrin puncta (new Fig. 5D).
(6) Quantification of protein numbers is challenging with SMLM. Issues include i) some of FP not correctly folded/mature, and ii) dependence of localisation rate on instrument, excitation/illumination intensities, and also the thresholds used in analysis. Can the authors compare with another protein that has known expression levels- e.g. PSD95? This is quite an ask, but if they could show copy number of something known to compare with, it would be useful.
We agree that absolute quantification with SMLM is challenging, since the number of detections depends on fluorophore maturation, photophysics, imaging conditions, and analysis thresholds (discussed in Patrizio & Specht 2016, Neurophotonics). For this reason, only very few datasets provide reliable copy numbers, even for well-studied proteins such as PSD-95. One notable exception is the study by Maynard et al. (eLife 2021) that quantified endogenous GlyRβcontaining receptors in spinal cord synapses using SMLM combined with correlative electron microscopy. The strength of this work was the use of a KI mouse strain, which ensures that mEos4b-GlyRβ expression follows intrinsic regional and temporal profiles. The authors reported a stereotypic density of ~2,000 GlyRs/µm² at synapses, corresponding to ~120 receptors per synapse in the dorsal horn and ~240 in the ventral horn, taking into account various parameters including receptor stoichiometry and the functionality of the fluorophore. These values are very close to our own calculations of GlyR numbers at spinal cord synapses that were obtained slightly differently in terms of sample preparation, microscope setup, imaging conditions, and data analysis, lending support to our experimental approach. Nevertheless, the obtained GlyR copy numbers at hippocampal synapses clearly have to be taken as estimates rather than precise figures, because the number of detections from a single mEos4b fluorophore can vary substantially, meaning that the fluorophores are not represented equally in pointillist images. This can affect the copy number calculation for a specific synapse, in particular when the numbers are low (e.g. in hippocampus), however, it should not alter the average number of detections (Fig. 1B) or the (median) molecule numbers of the entire population of synapses (Fig. 1C). We have discussed the limitations of our approach (p. 11).
(7) Rationale for doing nanobody dSTORM not clear at all. They don't explain the reason for doing the dSTORM experiments. Why not just rely on PALM for coincidence measurements, rather than tagging mEoS with a nanobody, and then doing dSTORM with that? Can they explain? Is it to get extra localisations- i.e. multiple per nanobody? If so, localising same FP multiple times wouldn't improve resolution. Also, no controls for nanobody dSTORM experiments- what about non-spec nb, or use on WT sections?
As discussed above (point 6), the detection of fluorophores with SMLM is influenced by many parameters, not least the noise produced by emitting molecules other than the fluorophore used for labelling. Our study is exceptional in that it attempts to identify extremely low molecule numbers (down to 1). To verify that the detections obtained with PALM correspond to mEos4b, we conducted robust control experiments (including pixel-shift as suggested by the reviewer, see point 1, revised Fig. 1B). The rationale for the nanobody-based dSTORM experiments was twofold: (1) to have an independent readout of the presence of low-copy GlyRs at inhibitory synapses and (2) to analyse the nanoscale organisation of GlyRs relative to the synaptic gephyrin scaffold using dual-colour dSTORM with spectral demixing (see p. 6). The organic fluorophores used in dSTORM (AF647, CF680) ensure high photon counts, essential for reliable co-localisation and distance analysis. PALM and dSTORM cannot be combined in dual-colour mode, as they require different buffers and imaging conditions.
The specificity of the anti-Eos nanobody was demonstrated by immunohistochemistry in spinal cord cultures expressing mEos4b-GlyRb and wildtype control tissue (Fig. S3). In response to the reviewer's remarks, we also performed a negative control experiment in Glrb<sup>eos/eos</sup> slices (dSTORM), in which the nanobody was omitted (new Fig. S4F,G). Under these conditions, spectral demixing produced a single peak corresponding to CF680 (gephyrin) without any AF647 contribution (Fig. S4F). The background detection of "false" AF647 detections at synapses was significantly lower than in the slices labelled with the nanobody. We conclude that the fluorescence signal observed in our dual-colour dSTORM experiments arises from the specific detection of mEos4b-GlyRb by the nanobody, rather than from background, crossreactivity or wrong attribution of colour during spectral demixing. We have added these data and explanations in the results (p. 7) and in the figure legend of Fig. S4F,G.
(8) What resolutions/precisions were obtained in SMLM experiments? Should perform Fourier Ring Correlation (FRC) on SR images to state resolutions obtained (particularly useful for when they're presenting distance histograms, as this will be dependent on resolution). Likewise for precision, what was mean precision? Can they show histograms of localisation precision.
This is an interesting question in the context of our experiments with low-copy GlyRs, since the spatial resolution of SMLM is limited also by the density of molecules, i.e. the sampling of the structure in question (Nyquist-Shannon criterion). Accordingly, the priority of the PALM experiments was to improve the sensibility of SMLM for the identification of mEos4b-GlyRb subunits, rather than to maximize the spatial resolution. The mean localisation precision in PALM was 33 +/- 12 nm, as calculated from the fitting parameters of each detection (Zeiss, ZEN software), which ultimately result from their signal-to-noise ratio. This is a relatively low precision for SMLM, which can be explained by the low brightness of mEos4b compared to organic fluorophores together with the elevated fluorescence background in tissue slices.
In the case of dSTORM, the aim was to study the relative distribution of GlyRs within the synaptic scaffold, for which a higher localisation precision was required (p. 6). Therefore, detections with a precision ≥ 25 nm were filtered during analysis with NEO software (Abbelight). The retained detections had a mean localisation precision of 12 +/- 5 for CF680 (Sylite) and 11 +/- 4 for AF647 (nanobody). These values are given in the revised manuscript (pp. 18, 22).
(9) Why were DBSCAN parameters selected? How can they rule out multiple localisations per fluor? If low copy numbers (<10), then why bother with DBSCAN? Could just measure distance to each one.
Multiple detections of the same fluorophore are intrinsic to dSTORM imaging and have not been eliminated from the analysis. Small clusters of detections likely represent individual molecules (e.g. single receptors in the extrasynaptic regions, Fig. 2A). DBSCAN is a robust clustering method that is quite insensitive to minor changes in the choice of parameters. For dSTORM of synaptic gephyrin clusters (CF680), a relatively low length (80 nm radius) together with a high number of detections (≥ 50 neighbours) were chosen to reconstruct the postsynaptic domain with high spatial resolution (see point 8). In the case of the GlyR (nanobody-AF647), the clustering was done mostly for practical reasons, as it provided the coordinates of the centre of mass of the detections. The low stringency of this clustering (200 nm radius, ≥ 5 neighbours) effectively filters single detections that can result from background noise or incorrect demixing. An additional reference explaining the use of DBSCAN including the choice of parameters is given on p. 22 (see also R2 point 4).
(10) For microscopy experiment methods, state power densities, not % or "nominal power".
Done. We now report the irradiance (laser power density) instead of nominal power (pp. 18, 21).
(11) In general, not much data presented. Any SI file with extra images etc.?
The original submission included four supplementary figures with additional data and representative images that should have been available to the reviewer (Figs. S1-S4). The SI file has been updated during revision (new Fig. S4E-G).
(12) Clarification of the discussion on GlyR expression and synaptic localization: The discussion on GlyR expression, complex formation, and synaptic localization is sometimes unclear, and needs terminological distinctions between "expression level", "complex formation" and "synaptic localization". For example, the authors state:"What then is the reason for the low protein expression of GlyRβ? One possibility is that the assembly of mature heteropentameric GlyR complexes depends critically on the expression of endogenous GlyR α subunits." Does this mean that GlyRβ proteins that fail to form complexes with GlyRα subunits are unstable and subject to rapid degradation? If so, the authors should clarify this point. The statement "This raises the interesting possibility that synaptic GlyRs may depend specifically on the concomitant expression of both α1 and β transcripts." suggests a dependency on α1 and β transcripts. However, is the authors' focus on synaptic localization or overall protein expression levels? If this means synaptic localization, it would be beneficial to state this explicitly to avoid confusion. To improve clarity, the authors should carefully distinguish between these different aspects of GlyR biology throughout the discussion. Additionally, a schematic diagram illustrating these processes would be highly beneficial for readers.
We thank the reviewer to point this out. We are dealing with several processes; protein expression that determines subunit availability and the assembly of pentameric GlyRs complexes, surface expression, membrane diffusion and accumulation of GlyRb-containing receptor complexes at inhibitory synapses. We have edited the manuscript, particularly the discussion and tried to be as clear as possible in our wording.
We chose not to add a schematic illustration for the time being, because any graphical representation is necessarily a simplification. Instead, we preferred to summarise the main numbers in tabular form (Table 1). We are of course open to any other suggestions.
(13) Interpretation of GlyR localization in the context of nanodomains. The distribution of GlyR molecules on inhibitory synapses appears to be non-homogeneous, instead forming nanoclusters or nanodomains, similar to many other synaptic proteins. It is important to interpret GlyR localization in the context of nanodomain organization.
The dSTORM images in Fig. 2 are pointillist representations that show individual detections rather than molecules. Small clusters of detections are likely to originate from a single AF647 fluorophore (in the case of nanobody labelling) and therefore represent single GlyRb subunits. Since GlyR copy numbers are so low at hippocampal synapses (≤ 5), the notion of nanodomain is not directly applicable. Our analysis therefore focused on the integration of GlyRs within the postsynaptic scaffold, rather than attempting to define nanodomain structures (see also response to point 8 of R1). A clarification has been added in the revised manuscript (p. 6).
Reviewer #1 (Significance):
The paper presents biological and technical advances. The biological insights revolve mostly on the documentation of Glycine receptors in particular synapses in forebrain, where they are typically expressed at very low levels. The authors provide compelling data indicating that the expression is of physiological significance. The authors have done a nice job of combining genetically-tagged mice with advanced microscopy methods to tackle the question of distributions of synaptic proteins. Overall these advances are more incremental than groundbreaking.
We thank the reviewer for acknowledging both the technical and biological advances of our study. While we recognize that our work builds upon established models, we consider that it also addresses important unresolved questions, namely that GlyRs are present and specifically anchored at inhibitory synapses in telencephalic regions, such as the hippocampus and striatum. From a methodological point of view, our study demonstrates that SMLM can be applied not only for structural analysis of highly abundant proteins, but also to reliably detect proteins present at very low copy numbers. This ability to identify and quantify sparse molecule populations adds a new dimension to SMLM applications, which we believe increases the overall impact of our study beyond the field of synaptic neuroscience.
Reviewer #2 (Evidence, reproducibility and clarity):
In their manuscript "Single molecule counting detects low-copy glycine receptors in hippocampal and striatal synapses" Camuso and colleagues apply single molecule localization microscopy (SMLM) methods to visualize low copy numbers of GlyRs at inhibitory synapses in the hippocampal formation and the striatum. SMLM analysis revealed higher copy numbers in striatum compared to hippocampal inhibitory synapses. They further provide evidence that these low copy numbers are tightly linked to post-synaptic scaffolding protein gephyrin at inhibitory synapses. Their approach profits from the high sensitivity and resolution of SMLM and challenges the controversial view on the presence of GlyRs in these formations although there are reports (electrophysiology) on the presence of GlyRs in these particular brain regions. These new datasets in the current manuscript may certainly assist in understanding the complexity of fundamental building blocks of inhibitory synapses.
However I have some minor points that the authors may address for clarification:
(1) In Figure 1 the authors apply PALM imaging of mEos4b-GlyRß (knockin) and here the corresponding Sylite label seems to be recorded in widefield, it is not clearly stated in the figure legend if it is widefield or super-resolved. In Fig 1 A - is the scale bar 5 µm? Some Sylite spots appear to be sized around 1 µm, especially the brighter spots, but maybe this is due to the lower resolution of widefield imaging? Regarding the statistical comparison: what method was chosen to test for normality distribution, I think this point is missing in the methods section.
This is correct; the apparent size of the Sylite spots does not reflect the real size of the synaptic gephyrin domain due to the limited resolution of widefield imaging including the detection of outof-focus light. We have clarified in the legend of Fig. 1A that Sylite labelling was with classic epifluorescence microscopy. The scale bar in Fig. 1A corresponds to 5 µm. Since the data were not normally distributed, nonparametric tests (Kruskal- Wallis one-way ANOVA with Dunn’s multiple comparison test or Mann-Whitney U-test for pairwise comparisons) were used (p. 23).
Moreover I would appreciate a clarification and/or citation that the knockin model results in no structural and physiological changes at inhibitory synapses, I believe this model has been applied in previous studies and corresponding clarification can be provided.
The Glrbeos/eos mouse model has been described previously and does not exhibit any structural or physiological phenotypes (Maynard et al. 2021 eLife). The issue was also raised by reviewer R1 (point 5) and has been clarified in the revised manuscript (p. 4).
(2) In the next set of experiments the authors switch to demixing dSTORM experiments - an explanation why this is performed is missing in the text - I guess better resolution to perform more detailed distance measurements? For these experiments: which region of the hippocampus did the authors select, I cannot find this information in legend or main text.
Yes, the dSTORM experiments enable dual-colour structural analysis at high spatial resolution (see response to R1 point 7). An explanation has been added (p. 6).
(3) Regarding parameters of demixing experiments: the number of frames (10.000) seems quite low and the exposure time higher than expected for Alexa 647. Can the authors explain the reason for chosing these particular parameters (low expression profile of the target - so better separation?, less fluorophores on label and shorter collection time?) or is there a reference that can be cited? The laser power is given in the methods in percentage of maximal output power, but for better comparison and reproducibility I recommend to provide the values of a power meter (kW/cm2) as lasers may change their maximum output power during their lifetime.
Acquisition parameters (laser power, exposure time) for dSTORM were chosen to obtain a good localisation precision (~12 nm; see R1 point 8). The number of frames is adequate to obtain well sampled gephyrin scaffolds in the CF680 channel. In the case of the GlyR (nanobody-AF647), the concept of spatial resolution does not really apply due to the low number of targets (see R1, point 13). Power density (irradiance) values have now been given (pp. 18, 21).
(4) For analysis of subsynaptic distribution: how did the authors decide to choose the parameters in the NEO software for DBSCAN clustering - was a series of parameters tested to find optimal conditions and did the analysis start with an initial test if data is indeed clustered (K-ripley) or is there a reference in literature that can be provided?
DBSCAN parameters were optimised manually, by testing different values. Identification of dense and well-delimited gephyrin clusters (CF680) was achieved with a small radius and a high number of detections (80 nm, ≥ 50 neighbours), whereas filtering of low-density background in the AF647 channel (GlyRs) required less stringent parameters (200 nm, ≥ 5) due to the low number of target molecules. Similar parameters were used in a previous publication (Khayenko et al. 2022, Angewandte Chemie). The reference has been provided on p. 22 (see also R1 point 9).
(5) A conclusion/discussion of the results presented in Figure 5 is missing in the text/discussion.
This part of the manuscript has been completely overhauled. It includes new experimental data, quantification of the data (new Fig.5), as well as the discussion and interpretation of our findings (see also R1, point 3). In agreement with our earlier interpretation, the data confirm that low availability of GlyRa1 subunits limits the expression and synaptic targeting of GlyRa1/b heteropentamers. The observation that GlyRa1 overexpression with lentivirus increases the size of the postsynaptic gephyrin domain further points to a structural role, whereby GlyRs can enhance the stability (and size) of inhibitory synapses in hippocampal neurons, even at low copy numbers (pp. 13-14).
(6) In line 552 "suspension" is misleading, better use "solution"
Done.
Reviewer #2 (Significance):
Significance: The manuscript provides new insights to presence of low-copy numbers by visualizing them via SMLM. This is the first report that visualizes GlyR optically in the brain applying the knock-in model of mEOS4b tagged GlyRß and quantifies their copy number comparing distribution and amount of GlyRs from hippocampus and striatum. Imaging data correspond well to electrophysiological measurements in the manuscript.
Field of expertise: Super-Resolution Imaging and corresponding analysis
Reviewer #4 (Evidence, reproducibility and clarity):
In this study, Camuso et al., make use of a knock-in mouse model expressing endogenously mEos4b-tagged GlyRβ to detect endogenous glycine receptors using single-molecule localization microscopy. The main conclusion from this study is that in the hippocampus GlyRβ molecules are barely detected, while inhibitory synapses in the ventral striatum seem to express functionally relevant GlyR numbers.
I have a few points that I hope help to improve the strength of this study.
- In the hippocampus, this study finds that the numbers of detections are very low. The authors perform adequate controls to indicate that these localizations are above noise level. Nevertheless, it remains questionable that these reflect proper GlyRs. The suggestion that in hippocampal synapses the low numbers of GlyRβ molecules "are important in assembly or maintenance of inhibitory synaptic structures in the brain" is on itself interesting, but is not at all supported. It is also difficult to envision how such low numbers could support the structure of a synapse. A functional experiment showing that knockdown of GlyRs affects inhibitory synapse structure in hippocampal neurons would be a minimal test of this.
It is not clear what the reviewer means by “it remains questionable that these reflect proper GlyRs”. The PALM experiments include a series of stringent controls (see R1, point 1) demonstrating the existence of low-copy GlyRs at inhibitory synapses in the hippocampus (Fig. 1) and in the striatum (Fig. 3), and are backed up by dSTORM experiments (Fig. 2). We have no reason to doubt that these receptors are fully functional (as demonstrated for the ventral striatum (Fig. 4). However, due to their low number, a role in inhibitory synaptic transmission is clearly limited, at least in the hippocampus and dorsal striatum.
We therefore propose a structural role, where the GlyRs could be required to stabilise the postsynaptic gephyrin domain in hippocampal neurons. This is based on the idea that the GlyRgephyrin affinity is much higher than that of the GABAAR-gephyrin interaction (reviewed in Kasaragod & Schindelin 2018 Front Mol Neurosci). Accordingly, there is a close relationship between GlyRs and gephyrin numbers, sub-synaptic distribution, and dynamics in spinal cord synapses that are mostly glycinergic (Specht et al. 2013 Neuron; Maynard et al. 2021 eLife; Chapdelaine et al. 2021 Biophys J). It is reasonable to assume that low-copy GlyRs could play a similar structural role at hippocampal synapses. A knockdown experiment targeting these few receptors is technically very challenging and beyond the scope of this study. However, in response to the reviewer's question we have conducted new experiments in cultured hippocampal neurons (new Fig. 5). They demonstrate that overexpression of GlyRa1/b heteropentamers increases the size of the postsynaptic domain in these neurons, supporting our interpretation of a structural role of low-copy GlyRs (p. 14).
- The endogenous tagging strategy is a very strong aspect of this study and provides confidence in the labeling of GlyRβ molecules. One caveat however, is that this labeling strategy does not discriminate whether GlyRβ molecules are on the cell membrane or in internal compartments. Can the authors provide an estimate of the ratio of surface to internal GlyRβ molecules?
Gephyrin is known to form a two-dimensional scaffold below the synaptic membrane to which inhibitory GlyRs and GABAARs attach (reviewed in Alvarez 2017 Brain Res). The majority of the synaptic receptors are therefore thought to be located in the synaptic membrane, which is supported by the close relationship between the sub-synaptic distribution of GlyRs and gephyrin in spinal cord neurons (e.g. Maynard et al. 2021 eLife). To demonstrate the surface expression of GlyRs at hippocampal synapses we labelled cultured hippocampal neurons expressing mEos4b-GlyRa1 with anti-Eos nanobody in non-permeabilised neurons (see Author response image 1). The close correspondence between the nanobody (AF647) and the mEos4b signal confirms that the majority of the GlyRs are indeed located in the synaptic membrane.
Author response image 1.
Left: Lentivirus expression of mEos4b-GlyRa1 in fixed and non-permeabilised hippocampal neurons (mEos4b signal). Right: Surface labelling of the recombinant subunit with anti-Eos nanoboby (AF647).
- “We also estimated the absolute number of GlyRs per synapse in the hippocampus. The number of mEos4b detections was converted into copy numbers by dividing the detections at synapses by the average number of detections of individual mEos4b-GlyRβ containing receptor complexes”. In essence this is a correct method to estimate copy numbers, and the authors discuss some of the pitfalls associated with this approach (i.e., maturation of fluorophore and detection limit). Nevertheless, the authors did not subtract the number of background localizations determined in the two negative control groups. This is critical, particularly at these low-number estimations.
We fully agree that background subtraction can be useful with low detection numbers. In the revised manuscript, copy numbers are now reported as background-corrected values. Specifically, the mean number of detections measured in wildtype slices was used to calculate an equivalent receptor number, which was then subtracted from the copy number estimates across hippocampus, spinal cord and striatum. This procedure is described in the methods (p. 20) and results (p. 5, 8), and mentioned in the figure legends of Fig. 1C, 3C. The background corrected values are given in the text and Table 1.
- Furthermore, the authors state that "The advantage of this estimation is that it is independent of the stoichiometry of heteropentameric GlyRs". However, if the stoichometry is unknown, the number of counted GlyRβ subunits cannot simply be reported as the number of GlyRs. This should be discussed in more detail, and more carefully reported throughout the manuscript.
The reviewer is right to point this out. There is still some debate about the stoichiometry of heteropentameric GlyRs. Configurations with 2a:3b, 3a:2b and 4a:1b subunits have been advanced (e.g. Grudzinska et al. 2005 Neuron; Durisic et al. 2012 J Neurosci; Patrizio et al. 2017 Sci Rep; Zhu & Gouaux 2021 Nature). We have therefore chosen a quantification that is independent of the underlying stoichiometry. Since our quantification is based on very sparse clusters of mEos4b detections that likely originate from a single receptor complex (irrespective of its stoichiometry), the reported values actually reflect the number of GlyRs (and not GlyRb subunits). We have clarified this in the results (p. 5) and throughout the manuscript (Table 1).
- The dual-color imaging provides insights in the subsynaptic distribution of GlyRβ molecules in hippocampal synapses. Why are similar studies not performed on synapses in the ventral striatum where functionally relevant numbers of GlyRβ molecules are found? Here insights in the subsynaptic receptor distribution would be of much more interest as it can be tight to the function.
This is an interesting suggestion. However, the primary aim of our study was to identify the existence of GlyRs in hippocampal regions. At low copy numbers, the concept of sub-synaptic domains (SSDs, e.g. Yang et al. 2021 EMBO Rep) becomes irrelevant (see R1 point 13). It should be pointed out that the dSTORM pointillist images (Fig. 2A) represent individual GlyR detections rather than clusters of molecules. In the striatum, our specific purpose was to solve an open question about the presence of GlyRs in different subregions (putamen, nucleus accumbens).
- It is unclear how the experiments in Figure 5 add to this study. These results are valid, but do not seem to directly test the hypothesis that "the expression of α subunits may be limiting factor controlling the number of synaptic GlyRs". These experiments simply test if overexpressed α subunits can be detected. If the α subunits are limiting, measuring the effect of α subunit overexpression on GlyRβ surface expression would be a more direct test.
Both R1 and R2 have also commented on the data in Fig. 5 and their interpretation. We have substantially revised this section as described before (see R1 point 3) including additional experiments and quantification of the data (new Fig. 5). The findings lend support to our earlier hypothesis that GlyR alpha subunits (in particular GlyRa1) are the limiting factor for the expression of heteropentameric GlyRa/b in hippocampal neurons (pp. 13-14). Since the GlyRa1 subunit itself does not bind to gephyrin (Patrizio et al. 2017 Sci Rep), the synaptic localisation of the recombinant mEos4b-GlyRa1 subunits is proof that they have formed heteropentamers with endogenous GlyRb subunits and driven their membrane trafficking, which the GlyRb subunits are incapable of doing on their own.
Reviewer #4 (Significance):
These results are based on carefully performed single-molecule localization experiments, and are well-presented and described. The knockin mouse with endogenously tagged GlyRβ molecules is a very strong aspect of this study and provides confidence in the labeling, the combination with single-molecule localization microscopy is very strong as it provides high sensitivity and spatial resolution.
The conceptual innovation however seems relatively modest, these results confirm previous studies but do not seem to add novel insights. This study is entirely descriptive and does not bring new mechanistic insights.
This study could be of interest to a specialized audience interested in glycine receptor biology, inhibitory synapse biology and super-resolution microscopy.
My expertise is in super-resolution microscopy, synaptic transmission and plasticity
As we have stated before, the novelty of our study lies in the use of SMLM for the identification of very small numbers of molecules, which requires careful control experiments. This is something that has not been done before and that can be of interest to a wider readership, as it opens up SMLM for ultrasensitive detection of rare molecular events. Using this approach, we solve two open scientific questions: (1) the demonstration that low-copy GlyRs are present at inhibitory synapses in the hippocampus, (2) the sub-region specific expression and functional role of GlyRs in the ventral versus dorsal striatum.
The following review was provided later under the name “Reviewer #4”. To avoid confusion with the last reviewer from above we will refer to this review as R4-2.
Reviewer #4-2 (Evidence, reproducibility and clarity):
Summary:
Provide a short summary of the findings and key conclusions (including methodology and model system(s) where appropriate).
The authors investigate the presence of synaptic glycine receptors in the telencephalon, whose presence and function is poorly understood.
Using a transgenically labeled glycine receptor beta subunit (Glrb-mEos4b) mouse model together with super-resolution microscopy (SLMM, dSTORM), they demonstrate the presence of a low but detectable amount of synaptically localized GLRB in the hippocampus. While they do not perform a functional analysis of these receptors, they do demonstrate that these subunits are integrated into the inhibitory postsynaptic density (iPSD) as labeled by the scaffold protein gephyrin. These findings demonstrate that a low level of synaptically localized glycerine receptor subunits exist in the hippocampal formation, although whether or not they have a functional relevance remains unknown.
They then proceed to quantify synaptic glycine receptors in the striatum, demonstrating that the ventral striatum has a significantly higher amount of GLRB co-localized with gephyrin than the dorsal striatum or the hippocampus. They then recorded pharmacologically isolated glycinergic miniature inhibitory postsynaptic currents (mIPSCs) from striatal neurons. In line with their structural observations, these recordings confirmed the presence of synaptic glycinergic signaling in the ventral striatum, and an almost complete absence in the dorsal striatum. Together, these findings demonstrate that synaptic glycine receptors in the ventral striatum are present and functional, while an important contribution to dorsal striatal activity is less likely.
Lastly, the authors use existing mRNA and protein datasets to show that the expression level of GLRA1 across the brain positively correlates with the presence of synaptic GLRB.
The authors use lentiviral expression of mEos4b-tagged glycine receptor alpha1, alpha2, and beta subunits (GLRA1, GLRA1, GLRB) in cultured hippocampal neurons to investigate the ability of these subunits to cause the synaptic localization of glycine receptors. They suggest that the alpha1 subunit has a higher propensity to localize at the inhibitory postsynapse (labeled via gephyrin) than the alpha2 or beta subunits, and may therefore contribute to the distribution of functional synaptic glycine receptors across the brain.
Major comments:
- Are the key conclusions convincing?
The authors are generally precise in the formulation of their conclusions.
(1) They demonstrate a very low, but detectable, amount of a synaptically localized glycine receptor subunit in a transgenic (GlrB-mEos4b) mouse model. They demonstrate that the GLRB-mEos4b fusion protein is integrated into the iPSD as determined by gephyrin labelling. The authors do not perform functional tests of these receptors and do not state any such conclusions.
(2) The authors show that GLRB-mEos4b is clearly detectable in the striatum and integrated into gephyrin clusters at a significantly higher rate in the ventral striatum compared to the dorsal striatum, which is in line with previous studies.
(3) Adding to their quantification of GLRB-mEos4b in the striatum, the authors demonstrate the presence of glycinergic miniature IPSCs in the ventral striatum, and an almost complete absence of mIPSCs in the dorsal striatum. These currents support the observation that GLRB-mEos4b is more synaptically integrated in the ventral striatum compared to the dorsal striatum.
(4) The authors show that lentiviral expression of GLRA1-mEos4b leads to a visually higher number of GLR clusters in cultured hippocampal neurons, and a co-localization of some clusters with gephyrin. The authors claim that this supports the idea that GLRA1 may be an important driver of synaptic glycine receptor localization. However, no quantification or statistical analysis of the number of puncta or their colocalization with gephyrin is provided for any of the expressed subunits. Such a claim should be supported by quantification and statistics
A thorough analysis and quantification of the data in Fig.5 has been carried out as requested by all the other reviewers (e.g. R1, point 3). The new data and results have been described in the revised manuscript (pp. 9-10, 13-14).
- Should the authors qualify some of their claims as preliminary or speculative, or remove them altogether?
One unaddressed caveat is the fact that a GLRB-mEos4b fusion protein may behave differently in terms of localization and synaptic integration than wild-type GLRB. While unlikely, it is possible that mEos4b interacts either with itself or synaptic proteins in a way that changes the fused GLRB subunit’s localization. Such an effect would be unlikely to affect synaptic function in a measurable way, but might be detected at a structural level by highly sensitive methods such as SMLM and STORM in regions with very low molecule numbers (such as the hippocampus). Since reliable antibodies against GLRB in brain tissue sections are not available, this would be difficult to test. Considering that no functional measures of the hippocampal detections exist, we would suggest that this possible caveat be mentioned for this particular experiment.
This question has also been raised before (R1, point 5). According to an earlier study the mEos4b-GlyRb knock-in does not cause any obvious phenotypes, with the possible exception of minor loss of glycine potency (Maynard et al. 2021 eLife). The fact that the synaptic levels in the spinal cord in heterozygous animals are precisely half of those of homozygous animals argues against differences in receptor expression, heteropentameric assembly, forward trafficking to the plasma membrane and integration into the synaptic membrane as confirmed using quantitative super-resolution CLEM (Maynard et al. 2021 eLife). Accordingly, we did not observe any behavioural deficits in these animals, making it a powerful experimental model. We have added this information in the revised manuscript (p. 4).
In addition, without any quantification or statistical analysis, the author’s claims regarding the necessity of GLRA1 expression for the synaptic localization of glycine receptors in cultured hippocampal neurons should probably be described as preliminary (Fig. 5).
As mentioned before, we have substantially revised this part (R1, point 3). The quantification and analysis in the new Fig. 5 support our earlier interpretation.
- Would additional experiments be essential to support the claims of the paper? Request additional experiments only where necessary for the paper as it is, and do not ask authors to open new lines of experimentation.
The authors show that there is colocalization of gephyrin with the mEos4b-GlyRβ subunit using the Dual-colour SMLM. This is a powerful approach that allows for a claim to be made on the synaptic location of the glycine receptors. The images presented in Figure 1, together with the distance analysis in Figure 2, display the co-localization of the fluorophores. The co-localization images in all the selected regions, hippocampus and striatum, also show detections outside of the gephyrin clusters, which the authors refer to as extrasynaptic. These punctated small clusters seem to have the same size as the ones detected and assigned as part of the synapse. It would be informative if the authors analysed the distribution, density and size of these nonsynaptic clusters and presented the data in the manuscript and also compared it against the synaptic ones. Validating this extrasynaptic signal by staining for a dendritic marker, such as MAP-2 or maybe a somatic marker and assessing the co-localization with the non-synaptic clusters would also add even more credibility to them being extrasynaptic.
The existence of extrasynaptic GlyRs is well attested in spinal cord neurons (e.g. Specht et al. 2013 Neuron; this study see Fig. S2). The fact that these appear as small clusters of detections in SMLM recordings results from the fact that a single fluorophore can be detected several times in consecutive image frames and because of blinking. Therefore, small clusters of detections likely represent single GlyRs (that can be counted), and not assemblies of several receptor complexes. Due to their diffusion in the neuronal membrane, they are seen as diffuse signals throughout the somatodendritic compartment in epifluorescence images (e.g. Fig. 5A). SMLM recordings of the same cells resolves this diffuse signal into discrete nanoclusters representing individual receptors (Fig. 5B). It is not clear what information co-localisation experiments with specific markers could provide, especially in hippocampal neurons, in which the copy numbers (and density) of GlyRs is next to zero.
In addition we would encourage the authors to quantify the clustering and co-localization of virally expressed GLRA1, GLRA2, and GLRB with gephyrin in order to support the associated claims (Fig. 5). Preferably, the density of GLR and gephyrin clusters (at least on the somatic surface, the proximal dendrites, or both) as well as their co-localization probability should be quantified if a causal claim about subunit-specific requirements for synaptic localization is to be made.
Quantification of the data have been carried out (new Fig.5C,D). The results have been described before (R1, point 3) and support our earlier interpretation of the data (pp. 13-14).
Lastly, even though it may be outside of the scope of such a study analysing other parts of the hippocampal area could provide additional important information. If one looks at the Allen Institute’s ISH of the beta subunit the strongest signal comes from the stratum oriens in the CA1 for example, suggesting that interneurons residing there would more likely have a higher expression of the glycine receptors. This could also be assessed by looking more carefully at the single cell transcriptomics, to see which cell types in the hippocampus show the highest mRNA levels. If the authors think that this is too much additional work, then perhaps a mention of this in the discussion would be good.
We have added the requested information from the ISH database of the Allen Institute in the discussion as suggested by the reviewer (p. 12). However, in combination with the transcriptomic data (Fig. S1) our finding strongly suggest that the expression of synaptic GlyRs depends on the availability of alpha subunits rather than on the presence of the GlyRb transcript. This is obvious when one compares the mRNA levels in the hippocampus with those in the basal ganglia (striatum) and medulla. While the transcript concentrations of GlyRb are elevated in all three regions and essentially the same, our data show that the GlyRb copy numbers at synapses differ over more than 2 orders of magnitude (Fig. 1B, Table 1).
- Are the suggested experiments realistic in terms of time and resources? It would help if you could add an estimated cost and time investment for substantial experiments.
Since the labeling and some imaging has been performed already, the requested experiment would be a matter of deploying a method of quantification. In principle, it should not require any additional wet-lab experiments, although it may require additional imaging of existing samples.
- Are the data and the methods presented in such a way that they can be reproduced?
Yes, for the most part.
- Are the experiments adequately replicated and statistical analysis adequate?
Yes
Minor comments:
- Specific experimental issues that are easily addressable.
N/A
- Are prior studies referenced appropriately?
Yes
- Are the text and figures clear and accurate?
Yes, although quantification in figure 5 is currently not present.
A quantification has been added (see R1, point 3).
- Do you have suggestions that would help the authors improve the presentation of their data and conclusions?
This paper presents a method that could be used to localize receptors and perhaps other proteins that are in low abundance or for which a detailed quantification is necessary. I would therefore suggest that Figure S4 is included into Figure 2 as the first panel, showcasing the demixing, followed by the results.
We agree in principle with this suggestion. However, the revised Fig. S4 is more complex and we think that it would distract from the data shown in Fig. 2. Given that Fig. S4 is mostly methodological and not essential to understand the text, we have kept it in the supplement for the time being. We leave the final decision on this point to the editor.
Reviewer #4-2 (Significance):
[This review was supplied later]
- Describe the nature and significance of the advance (e.g. conceptual, technical, clinical) for the field.
Using a novel and high resolution method, the authors have provided strong evidence for the presence of glycine receptors in the murine hippocampus and in the dorsal striatum. The number of receptors calculated is small compared to the numbers found in the ventral striatum. This is the first study to quantify receptor numbers in these region. In addition it also lays a roadmap for future studies addressing similar questions.
- Place the work in the context of the existing literature (provide references, where appropriate).
This is done well by the authors in the curation of the literature. As stated above, the authors have filled a gap in the presence of glycine receptors in different brain regions, a subject of importance in understanding the role they play in brain activity and function.
- State what audience might be interested in and influenced by the reported findings.
Neuroscientists working at the synaptic level, on inhibitory neurotransmission and on fundamental mechanisms of expression of genes at low levels and their relationship to the presence of the protein would be interested. Furthermore, researchers in neuroscience and cell biology may benefit from and be inspired by the approach used in this manuscript, to potentially apply it to address their own aims.
We thank the reviewer for the positive assessment of the technical and biological implications of our work, as well as the interest of our findings to a wide readership of neuroscientists and cell biologists.
- Define your field of expertise with a few keywords to help the authors contextualize your point of view. Indicate if there are any parts of the paper that you do not have sufficient expertise to evaluate.
Synaptic transmission, inhibitory cells and GABAergic synapses functionally and structurally, cortex and cortical circuits. No strong expertise in super-resolution imaging methods.
-
-
scottbevill.net scottbevill.net
-
Do not be jealous of your sister. Know that diamonds and roses are as uncomfortable when they tumble from one’s lips as toads and frogs: colder, too, and sharper, and they cut.
There were a couple moments in here that I think are talking about other fairy tales and fables, but this is the only one where I can remember the specific story that it's talking about— and I wonder if every instruction is a direct reference to another story or if they're just so seamlessly interwoven that I can't tell which ones are exclusive to this one
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the current reviews.
We would like to thank the reviewers for their efforts and feedback on our preprint. We have elected to rework the manuscript for publication in a different journal. In this process we will alter many of the approaches and re-evaluate the conclusions. With this, many of the points raised by the reviewers will be no longer relevant and therefore do not require a response. Again, we thank the reviewers for their time and helpful feedback.
The following is the authors’ response to the original reviews.
eLife Assessment:
The authors present a potentially useful approach of broad interest arguing that anterior cingulate cortex (ACC) tracks option values in decisions involving delayed rewards. The authors introduce the idea of a resource-based cognitive effort signal in ACC ensembles and link ACC theta oscillations to a resistance-based strategy. The evidence supporting these new ideas is incomplete and would benefit from additional detail and more rigorous analyses and computational methods.
We are extremely grateful for the several excellent and comments of the reviewers. To address these concerns, we have completely reworked the manuscript adding more rigorous approaches in each phase of the analysis and computational model. We realize that this has taken some time to prepare the revision. However, given the comments of the reviewers, we felt it necessary to thoroughly rework the paper based on their input. Here is a (nonexhaustive) overview of the major changes we made:
We have developed a way to more adequately capture the heterogeneity in the behavior
We have completely reworked the RL model
We have added additional approaches and rigor to the analysis of the value-tracking signal.
Reviewer #1 (Public Review):
Summary:
Young (2.5 mo [adolescent]) rats were tasked to either press one lever for immediate reward or another for delayed reward.
Please note that at the time of testing and training that the rats were > 4 months old.
The task had a complex structure in which (1) the number of pellets provided on the immediate reward lever changed as a function of the decisions made, (2) rats were prevented from pressing the same lever three times in a row. Importantly, this task is very different from most intertemporal choice tasks which adjust delay (to the delayed lever), whereas this task held the delay constant and adjusted the number of 20 mg sucrose pellets provided on the immediate value lever.
Several studies parametrically vary the immediate lever (PMID: 39119916, 31654652, 28000083, 26779747, 12270518, 19389183). While most versions of the task will yield qualitatively similar estimates of discounting, the adjusting amount is preferred as it provides the most consistent estimates (PMID: 22445576). More specifically this version of the task avoids contrast effects of that result from changing the delay during the session (PMID: 23963529, 24780379, 19730365, 35661751) which complicates value estimates.
Analyses are based on separating sessions into groups, but group membership includes arbitrary requirements and many sessions have been dropped from the analyses.
We have updated this approach and now provide a more comprehensive assessment of the behavior. The updated approach applies a hierarchical clustering model to the behavior in each session. This was applied at each delay to separate animals that prefer the immediate option more/less. This results in 4 statistically dissociable groups (4LO, 4HI, 8LO, 8HI) and includes all sessions. Please see Figure 1.
Computational modeling is based on an overly simple reinforcement learning model, as evidenced by fit parameters pegging to the extremes.
We have completely reworked the simulations in the revision. In the updated RL model we carefully add parameters to determine which are necessary to explain the experimental data. We feel that it is simplified yet more descriptive. Please see Figure 2 and associated text.
The neural analysis is overly complex and does not contain the necessary statistics to assess the validity of their claims.
We have dramatically streamlined the spike train analysis approach and added several statistical tests to ensure the rigor of our results. Please see Figures 4,5,6 and associated text.
Strengths:
The task is interesting.
Thank you for the positive comment
Weaknesses:
Behavior:
The basic behavioral results from this task are not presented. For example, "each recording session consisted of 40 choice trials or 45 minutes". What was the distribution of choices over sessions? Did that change between rats? Did that change between delays? Were there any sequence effects? (I recommend looking at reaction times.) Were there any effects of pressing a lever twice vs after a forced trial?
Please see the updated statistics and panels in Figures 1 and 2. We believe these address this valid concern.
This task has a very complicated sequential structure that I think I would be hard pressed to follow if I were performing this task.
Human tasks implement a similar task structure (PMID: 26779747). Please note the response above that outlines the benefits of using of this task.
Before diving into the complex analyses assuming reinforcement learning paradigms or cognitive control, I would have liked to have understood the basic behaviors the rats were taking. For example, what was the typical rate of lever pressing? If the rats are pressing 40 times in 45 minutes, does waiting 8s make a large difference?
Thank you for this suggestion. Our additions to Figure 1 are intended to better explain and quantify the behavior of the animals. Note that this task is designed to hold the rate of reinforcement constant no matter the choices of the animals. Our analysis supports the long-held view in the literature that rats do not like waiting for rewards, even at small delays. Going from the 4 à 8 sec delay results in significantly more immediate choices, indicating that the rats will forgo waiting 8 sec for a larger reinforcer and take a smaller reinforcer at 4 sec.
For that matter, the reaction time from lever appearance to lever pressing would be very interesting (and important). Are they making a choice as soon as the levers appear? Are they leaning towards the delay side, but then give in and choose the immediate lever? What are the reaction time hazard distributions?
This is an excellent suggestion, we have added a brief analysis of reaction times (Please see the section entitled “4 behavioral groups are observed across all sessions” in the Results). Please note that an analysis of the reaction times has been presented in a prior analysis of this data set (White et al., 2024). In addition, an analysis of reaction times in this task was performed in Linsenbardt et al. (2017). In short, animals tend to choose within 1 second of the lever appearing. In addition, our prior work shows that responses on the immediate lever tend to be slower, which we viewed as evidence of increased deliberation requirements (possibly required to integrate value signals).
It is not clear that the animals on this task were actually using cognitive control strategies on this task. One cannot assume from the task that cognitive control is key. The authors only consider a very limited number of potential behaviors (an overly simple RL model). On this task, there are a lot of potential behavioral strategies: "win-stay/lose-shift", "perseveration", "alternation", even "random choices" should be considered.
The strategies the Reviewer mentioned are descriptors of the actual choices the rats made. For example, perseveration means the rat is choosing one of the levers at an excessively high rate whereas alternation means it is choosing the two levers more or less equally, independent of payouts. But the question we are interested in is why? We are arguing that the type of cognitive control determines the choice behavior, but cognitive control is an internal variable that guides behavior, rather than simply a descriptor of the behavior. For example, the animal opts to perseverate on the delayed lever because the cognitive control required to track ival is too high. We then searched the neural data for signatures of the two types of cognitive control.
The delay lever was assigned to the "non-preferred side". How did side bias affect the decisions made?
The side bias clearly does not impact performance as the animals prefer the delay lever at shorter delays, which works against this bias.
The analyses based on "group" are unjustified. The authors compare the proportion of delayed to immediate lever press choices on the non-forced trials and then did k-means clustering on this distribution. But the distribution itself was not shown, so it is unclear whether the "groups" were actually different. They used k=3, but do not describe how this arbitrary number was chosen. (Is 3 the optimal number of clusters to describe this distribution?) Moreover, they removed three group 1 sessions with an 8s delay and two group 2 sessions with a 4s delay, making all the group 1 sessions 4s delay sessions and all group 2 sessions 8s delay sessions. They then ignore group 3 completely. These analyses seem arbitrary and unnecessarily complex. I think they need to analyze the data by delay. (How do rats handle 4s delay sessions? How do rats handle 6s delay sessions? How do rats handle 8s delay sessions?). If they decide to analyze the data by strategy, then they should identify specific strategies, model those strategies, and do model comparison to identify the best explanatory strategy. Importantly, the groups were session-based, not rat based, suggesting that rats used different strategies based on the delay to the delayed lever.
We have completely reworked our approach for capturing the heterogeneity in behavior. We have taken care to show more of the behavioral statistics that have gone into identifying each of the groups. All sessions are included in this analysis. As the reviewer suggests, we used the statistics from each of the behavioral groups to inform the RL model that explores neural signals that underly decisions in this task. We strongly disagree that groups should be rat and not session based as the behavior of the animal can, and does, change from day to day. This is important to consider when analyzing the neural data as rat-based groupings would ignore this potential source of variance.
The reinforcement learning model used was overly simple. In particular, the RL model assumes that the subjects understand the task structure, but we know that even humans have trouble following complex task structures. Moreover, we know that rodent decision-making depends on much more complex strategies (model-based decisions, multi-state decisions, rate-based decisions, etc). There are lots of other ways to encode these decision variables, such as softmax with an inverse temperature rather than epsilon-greedy. The RL model was stated as a given and not justified. As one critical example, the RL model fit to the data assumed a constant exponential discounting function, but it is well-established that all animals, including rodents, use hyperbolic discounting in intertemporal choice tasks. Presumably this changes dramatically the effect of 4s and 8s. As evidence that the RL model is incomplete, the parameters found for the two groups were extreme. (Alpha=1 implies no history and only reacting to the most recent event. Epsilon=0.4 in an epsilongreedy algorithm is a 40% chance of responding randomly.)
While we agree that the approach was not fully justified, we do not agree that it was invalid. Simply stated, a softmax approach gives the best fit to the choice behavior, whereas our epsilon-greedy approach attempted to reproduce the choice behavior using a naïve agent that progressively learns the values of the two levers on a choice-by-choice basis. Nevertheless, we certainly appreciate that important insights can be gained by fitting a model to the data as suggested. We feel that the new modeling approach we have now implemented is optimal for the present purposes and it replaces the one used in the original manuscript.
The authors do add a "dbias" (which is a preference for the delayed lever) term to the RL model, but note that it has to be maximal in the 4s condition to reproduce group 2 behavior, which means they are not doing reinforcement learning anymore, just choosing the delayed lever.
The dbias term was dropped in the new model implementation
Neurophysiology:
The neurophysiology figures are unclear and mostly uninterpretable; they do not show variability, statistics or conclusive results.
While the reviewer is justified in criticizing the clarity of the figures, the statement that “they do not show variability, statistics or conclusive results” is not correct. Each of the figures presented in the first draft of the manuscript, except Figure 3, are accompanied by statistics and measures of variability. Nonetheless we have updated each of the neurophysiology analyses. We hope that the reviewer will find our updates more rigorous and thorough.
As with the behavior, I would have liked to have seen more traditional neurophysiological analyses first. What do the cells respond to? How do the manifolds change aligned to the lever presses? Are those different between lever presses?
We have added several figures that plot the mean +/- SEM of the neural activity (see Figures 4 and 5). Hopefully this provides a more intuitive picture of the changes in neural activity throughout the task.
Are there changes in cellular information (both at the individual and ensemble level) over time in the session?
We provide several analyses of how firing rate changes over trials in relation to ival over time and trials in the session. In addition, we describe how these signals change in each of the behavioral groups.
How do cellular responses differ during that delay while both levers are out, but the rats are not choosing the immediate lever?
We were somewhat unclear about this suggestion as the delay follows the lever press. In addition, there is no delay after immediate presses
Figure 3, for example, claims that some of the principal components tracked the number of pellets on the immediate lever ("ival"), but they are just two curves. No statistics, controls, or justification for this is shown. BTW, on Figure 3, what is the event at 200s?
This comment is no longer relevant based on the changes we’ve made to the manuscript.
I'm confused. On Figure 4, the number of trials seems to go up to 50, but in the methods, they say that rats received 40 trials or 45 minutes of experience.
This comment is no longer relevant based on the changes we’ve made to the manuscript.
At the end of page 14, the authors state that the strength of the correlation did not differ by group and that this was "predicted" by the RL modeling, but this statement is nonsensical, given that the RL modeling did not fit the data well, depended on extreme values. Moreover, this claim is dependent on "not statistically detectable", which is, of course, not interpretable as "not different".
This comment is no longer relevant based on the changes we’ve made to the manuscript.
There is an interesting result on page 16 that the increases in theta power were observed before a delayed lever press but not an immediate lever press, and then that the theta power declined after an immediate lever press.
Thank you for the positive comment.
These data are separated by session group (again group 1 is a subset of the 4s sessions, group 2 is a subset of the 8s sessions, and group 3 is ignored). I would much rather see these data analyzed by delay itself or by some sort of strategy fit across delays.
Thank you for the excellent suggestion. Our new group assignments take delay into account.
That being said, I don't see how this description shows up in Figure 6. What does Figure 6 look like if you just separate the sessions by delay?
We are unclear what the reviewer means by “this description”.
Discussion:
Finally, it is unclear to what extent this task actually gets at the questions originally laid out in the goals and returned to in the discussion. The idea of cognitive effort is interesting, but there is no data presented that this task is cognitive at all. The idea of a resourced cognitive effort and a resistance cognitive effort is interesting, but presumably the way one overcomes resistance is through resourcelimited components, so it is unclear that these two cognitive effort strategies are different.
The basis for the reviewers assertation that “the way one overcomes resistance is through resourcelimited components” is not clear. In the revised version, we have taken greater care to outline how each type of effort signal facilitates performance of the task and articulate these possibilities in our stochastic and RL models. We view the strong evidence for ival tracking presented herein as a critical component of resource based cognitive effort.
The authors state that "ival-tracking" (neurons and ensembles that presumably track the number of pellets being delivered on the immediate lever - a fancy name for "expectations") "taps into a resourced-based form of cognitive effort", but no evidence is actually provided that keeping track of the expectation of reward on the immediate lever depends on attention or mnemonic resources. They also state that a "dLP-biased strategy" (waiting out the delay) is a "resistance-based form of cognitive effort" but no evidence is made that going to the delayed side takes effort.
We challenge the reviewers that assertation ival tracking is a “fancy name for expectations”. We make no claim about the prospective or retrospective nature of the signal. Clearly, expectations should be prospective and therefore different from ival tracking. Regarding the resistance signal: First, animals avoid the delay lever more often at the 8 sec delay (Figure 1). We have shown that increasing the delay systematically biases responses AWAY from the delay (Linsenbardt et al., 2017). This is consistent with a well-developed literature that rats and mice do not like waiting for delayed reinforcers. We contend that enduring something you don’t like takes effort.
The authors talk about theta synchrony, but never actually measure theta synchrony, particularly across structures such as amygdala or ventral hippocampus. The authors try to connect this to "the unpleasantness of the delay", but provide no measures of pleasantness or unpleasantness. They have no evidence that waiting out an 8s delay is unpleasant.
We have added spike-field coherence to better contact the literature on synchrony. Note that we never refer to our results as “synchrony”. However, we would be remiss to not address the growing literature on theta synchrony in effort allocation. There is a well-developed literature that rats and mice do not like waiting for delayed reinforcers. If waiting out the delay was not pleasant then why do the animals forgo larger rewards to avoid it?
The authors hypothesize that the "ival-tracking signal" (the expectation of number of pellets on the immediate lever) "could simply reflect the emotional or autonomic response". Aside from the fact that no evidence for this is provided, if this were to be true, then, in what sense would any of these signals be related to cognitive control?
This is proposed as an alternative explanation to the ival signal in the discussion. It was added as our due diligence. Emotional state could provide feedback to the currently implemented control mechanism. If waiting for reinforcement is too unpleasant this could drive them to ival tracking and choosing the immediate option more frequently. We provide this option only as a possibility, not a conclusion. We have clarified this in the revised text. Nevertheless, based on our review of the literature, autonomic tracking in some form, seems to be the most likely function of ACC (Seamans & Floresco 2022). While the reviewer may disagree with this, we feel it is at least as valid as all the complex, cognitively-based interpretations that commonly appear in the literature.
Reviewer #2 (Public Review):
Summary:
This manuscript explores the neuronal signals that underlie resistance vs resource-based models of cognitive effort. The authors use a delayed discounting task and computational models to explore these ideas. The authors find that the ACC strongly tracks value and time, which is consistent with prior work. Novel contributions include quantification of a resource-based control signal among ACC ensembles, and linking ACC theta oscillations to a resistance-based strategy.
Strengths:
The experiments and analyses are well done and have the potential to generate an elegant explanatory framework for ACC neuronal activity. The inclusion of local-field potential / spike-field analyses is particularly important because these can be measured in humans.
Thank you for the endorsement of our work.
Weaknesses:
I had questions that might help me understand the task and details of neuronal analyses.
(1) The abstract, discussion, and introduction set up an opposition between resource and resistancebased forms of cognitive effort. It's clear that the authors find evidence for each (ACC ensembles = resource, theta=resistance?) but I'm not sure where the data fall on this dichotomy.
(a) An overall very simple schematic early in the paper (prior to the MCML model? or even the behavior) may help illustrate the main point.
(b) In the intro, results, and discussion, it may help to relate each point to this dichotomy.
(c) What would resource-based signals look like? What would resistance based signals look like? Is the main point that resistance-based strategies dominate when delays are short, but resource-based strategies dominate when delays are long?
(d) I wonder if these strategies can be illustrated? Could these two measures (dLP vs ival tracking) be plotted on separate axes or extremes, and behavior, neuronal data, LFP, and spectral relationships be shown on these axes? I think Figure 2 is working towards this. Could these be shown for each delay length? This way, as the evidence from behavior, model, single neurons, ensembles, and theta is presented, it can be related to this framework, and the reader can organize the findings.
These are excellent suggestions, and we have implemented them, where possible.
(2) The task is not clear to me.
(a) I wonder if a task schematic and a flow chart of training would help readers.
Yes, excellent idea, we have now included this in Figure 1.
(b) This task appears to be relatively new. Has it been used before in rats (Oberlin and Grahame is a mouse study)? Some history / context might help orient readers.
Indeed, this task has been used in rats in several prior studies in rats. Please see the following references (PMID: 39119916, 31654652, 28000083, 26779747, 12270518, 19389183).
(c) How many total sessions were completed with ascending delays? Was there criteria for surgeries? How many total recording sessions per animal (of the 54?)
Please note that the delay does not change within a session. There were no criteria for surgery.
(d) How many trials completed per session (40 trials OR 45 minutes)? Where are there errors? These details are important for interpreting Figure 1.
Every animal in this data set completed 40 trials and we have updated the task description to clarify this issue. There are no errors in this task, but rather the task is designed to the tendency to make an impulsive choice (smaller reward now).
(3) Figure 1 is unclear to me.
(a) Delayed vs immediate lever presses are being plotted - but I am not sure what is red, and what is blue. I might suggest plotting each animal.
We have updated Figure 1 considerably for clarity.
(b) How many animals and sessions go into each data point?
We hope this is clarified now with our new group assignments as all sessions were included in the analysis.
(c) Table 1 (which might be better referenced in the paper) refers to rats by session. Is it true that some rats (2 and 8) were not analyzed for the bulk of the paper? Some rats appear to switch strategies, and some stay in one strategy. How many neurons come from each rat?
We have updated Table 1 based on our new groupings. The rats that contribute the most sessions also tend to be represented across the behavioral groups therefore it is unlikely that effort allocation strategies across groupings are an esoteric feature of an animal.
(d) Task basics - RT, choice, accuracy, video stills - might help readers understand what is going into these plots
(e) Does the animal move differently (i.e., RTs) in G1 vs. G2?
Excellent suggestion. We have added more analysis of the task variables in the revision (e.g. RT, choice comparisons across delays, etc…)
(4) I wasn't sure how clustered G1 vs. G2 vs G3 are. To make this argument, the raw data (or some axis of it) might help.
(a) This is particularly important because G3 appears to be a mix of G1 and G2, although upon inspection, I'm not sure how different they really are
(b) Was there some objective clustering criteria that defined the clusters?
(c) Why discuss G3 at all? Can these sessions be removed from analysis?
Based on our updates to the behavioral analysis these comments are no longer relevant.
(5) The same applies to neuronal analyses in Fig 3 and 4
(a) What does a single neuron peri-event raster look like? I would include several of these.
(b) What does PC1, 2 and 3 look like for G1, G2, and G3?
(c) Certain PCs are selected, but I'm not sure how they were selected - was there a criteria used? How was the correlation between PCA and ival selected? What about PCs that don't correlate with ival?
(d) If the authors are using PCA, then scree plots and PETHs might be useful, as well as comparisons to PCs from time-shuffled / randomized data.
We hope that our reworking of the neural data analysis has clarified these issues. We now include several firing rate examples and aggregate data.
(6) I had questions about the spectral analysis
(a) Theta has many definitions - why did the authors use 6-12 Hz? Does it come from the hippocampal literature, and is this the best definition of theta? What about other bands (delta - 1-4 Hz), theta (4-7 Hz); and beta - 13- 30 Hz? These bands are of particular importance because they have been associated with errors, dopamine, and are abnormal in schizophrenia and Parkinson's disease.
This designation comes mainly from the hippocampal and ACC literature in rodents. In addition, this range best captured the peak in the power spectrum in our data. Note that we focus our analysis on theta give the literature regarding theta in the ACC as a correlate of cognitive controls (references in manuscript). We did interrogate other bands as a sanity check and the results were mostly limited to theta. Given the scope of our manuscript and the concerns raised regarding complexity we are concerned that adding frequency analyses beyond theta obfuscates the take home message.
However, the spectrograms in Figure 3 show a range of frequencies and highlight the ones in the theta band as the most dynamic prior to the choice.
(b) Power spectra and time-frequency analyses may justify the authors focus. I would show these (yaxis - frequency, x-axis - time, z-axis, power).
Thank you for the suggestion. We have added this to Figure 3.
(7) PC3 as an autocorrelation doesn't seem the to be right way to infer theta entrainment or spikefield relationships, as PCA can be vulnerable to phantom oscillations, and coherence can be transient. It is also difficult to compare to traditional measures of phase-locking. Why not simply use spike-field coherence? This is particularly important with reference to the human literature, which the authors invoke.
Excellent suggestion. Note that PCA provided a way to classify neurons that exhibited peaks in the autocorrelation at theta frequencies. We have added spike-field coherence, and this analysis confirms the differences in theta entrainment of the spike trains across the behavioral groups. Please see Figure 6D.
Reviewer #3 (Public Review):
Summary:
The study investigated decision making in rats choosing between small immediate rewards and larger delayed rewards, in a task design where the size of the immediate rewards decreased when this option was chosen and increased when it was not chosen. The authors conceptualise this task as involving two different types of cognitive effort; 'resistance-based' effort putatively needed to resist the smaller immediate reward, and 'resource-based' effort needed to track the changing value of the immediate reward option. They argue based on analyses of the behaviour, and computational modelling, that rats use different strategies in different sessions, with one strategy in which they consistently choose the delayed reward option irrespective of the current immediate reward size, and another strategy in which they preferentially choose the immediate reward option when the immediate reward size is large, and the delayed reward option when the immediate reward size is small. The authors recorded neural activity in anterior cingulate cortex (ACC) and argue that ACC neurons track the value of the immediate reward option irrespective of the strategy the rats are using. They further argue that the strategy the rats are using modulates their estimated value of the immediate reward option, and that oscillatory activity in the 6-12Hz theta band occurs when subjects use the 'resistancebased' strategy of choosing the delayed option irrespective of the current value of the immediate reward option. If solid, these findings will be of interest to researchers working on cognitive control and ACCs involvement in decision making. However, there are some issues with the experiment design, reporting, modelling and analysis which currently preclude high confidence in the validity of the conclusions.
Strengths:
The behavioural task used is interesting and the recording methods should enable the collection of good quality single unit and LFP electrophysiology data. The authors recorded from a sizable sample of subjects for this type of study. The approach of splitting the data into sessions where subjects used different strategies and then examining the neural correlates of each is in principle interesting, though I have some reservations about the strength of evidence for the existence of multiple strategies.
Thank you for the positive comments.
Weaknesses:
The dataset is very unbalanced in terms of both the number of sessions contributed by each subject, and their distribution across the different putative behavioural strategies (see table 1), with some subjects contributing 9 or 10 sessions and others only one session, and it is not clear from the text why this is the case. Further, only 3 subjects contribute any sessions to one of the behavioural strategies, while 7 contribute data to the other such that apparent differences in brain activity between the two strategies could in fact reflect differences between subjects, which could arise due to e.g. differences in electrode placement. To firm up the conclusion that neural activity is different in sessions where different strategies are thought to be employed, it would be important to account for potential cross-subject variation in the data. The current statistical methods don't do this as they all assume fixed effects (e.g. using trials or neurons as the experimental unit and ignoring which subject the neuron/trial came from).
In the revised manuscript we have updated the group assignments. We have improved our description of the logic and methods for employing these groupings as well. With this new approach, all sessions are now included in the analysis. The group assignments are made purely on the behavioral statistics of an animal in each session. We feel this approach is preferable to eliminating neurons or session with the goal of balancing them, which may introduce bias. Further, the rats that contribute the most sessions also tend to be represented across the behavioral groups therefore it is unlikely that effort allocation strategies across groupings are an esoteric feature of an animal. As neurons are randomly sampled from each animal on a given session, we feel that we’re justified in treating these as fixed effects.
It is not obvious that the differences in behaviour between the sessions characterised as using the 'G1' and 'G2' strategies actually imply the use of different strategies, because the behavioural task was different in these sessions, with a shorter wait (4 seconds vs 8 seconds) for the delayed reward in the G1 strategy sessions where the subjects consistently preferred the delayed reward irrespective of the current immediate reward size. Therefore the differences in behaviour could be driven by difference in the task (i.e. external world) rather than a difference in strategy (internal to the subject). It seems plausible that the higher value of the delayed reward option when the delay is shorter could account for the high probability of choosing this option irrespective of the current value of the immediate reward option, without appealing to the subjects using a different strategy.
Further, even if the differences in behaviour do reflect different behavioural strategies, it is not obvious that these correspond to allocation of different types of cognitive effort. For example, subjects' failure to modify their choice probabilities to track the changing value of the immediate reward option might be due simply to valuing the delayed reward option higher, rather than not allocating cognitive effort to tracking immediate option value (indeed this is suggested by the neural data). Conversely, if the rats assign higher value to the delayed reward option in the G1 sessions, it is not obvious that choosing it requires overcoming 'resistance' through cognitive effort.
The RL modelling used to characterise the subject's behavioural strategies made some unusual and arguably implausible assumptions:
Thank you for the feedback, based on these comments (and those above) we have completely reworked the RL model. In addition, we’ve taken care to separate out the variables that correspond to a resistance- versus a resource-based signal.
There were also some issues with the analyses of neural data which preclude strong confidence in their conclusions:
Figure 4I makes the striking claim that ACC neurons track the value of the immediately rewarding option equally accurately in sessions where two putative behavioural strategies were used, despite the behaviour being insensitive to this variable in the G1 strategy sessions. The analysis quantifies the strength of correlation between a component of the activity extracted using a decoding analysis and the value of the immediate reward option. However, as far as I could see this analysis was not done in a cross-validated manner (i.e. evaluating the correlation strength on test data that was not used for either training the MCML model or selecting which component to use for the correlation). As such, the chance level correlation will certainly be greater than 0, and it is not clear whether the observed correlations are greater than expected by chance.
We have added more rigorous methods to assess the ival tracking signal (Figure 4 and 5). In addition, we’ve dropped the claim that ival tracking is the same across the behavioral groups. We suspect that this was an artifact of a suboptimal group assignment approach in the previous version.
An additional caveat with the claim that ACC is tracking the value of the immediate reward option is that this value likely correlates with other behavioural variables, notably the current choice and recent choice history, that may be encoded in ACC. Encoding analyses (e.g. using linear regression to predict neural activity from behavioural variables) could allow quantification of the variance in ACC activity uniquely explained by option values after controlling for possible influence of other variables such as choice history (e.g. using a coefficient of partial determination).
We agree that the ival tracking signal may be influenced by other variables – especially ones that are not cognitive but rather more generated by the autonomic system. We have included a discussion of this possibility in the Discussion section. Our previous work has explored the role of choice history on neural activity, please see White et al., (2024).
Figure 5 argues that there are systematic differences in how ACC neurons represent the value of the immediate option (ival) in the G1 and G2 strategy sessions. This is interesting if true, but it appears possible that the effect is an artefact of the different distribution of option values between the two session types. Specifically, due to the way that ival is updated based on the subjects' choices, in G1 sessions where the subjects are mostly choosing the delayed option, ival will on average be higher than in G2 sessions where they are choosing the immediate option more often. The relative number of high, medium and low ival trials in the G1 and G2 sessions will therefore be different, which could drive systematic differences in the regression fit in the absence of real differences in the activity-value relationship. I have created an ipython notebook illustrating this, available at: https://notebooksharing.space/view/a3c4504aebe7ad3f075aafaabaf93102f2a28f8c189ab9176d48 07cf1565f4e3. To verify that this is not driving the effect it would be important to balance the number of trials at each ival level across sessions (e.g. by subsampling trials) before running the regression.
This is an excellent point and lead us to abandon the linear regression-based approach to quantify differences in ival coding across behavioral groups.
Recommendations for the authors:
Reviewer #1 (Recommendations For The Authors):
This paper was extremely hard to read. In addition to the issues raised in the public review (overly complex and incomplete analyses), one of the hardest things to deal with was the writing.
Thank you for the feedback. Hopefully we have addressed this with our thorough rewrite.
The presentation was extremely hard to follow. I had to read through it several times to figure out what the task was. It wasn't until I got to the RL model Figure 2A that I realized what was really going on with the task. I strongly recommend having an initial figure that lays out the actual task (without any RL or modeling assumptions) and identifies the multiple different kinds of sessions. What is the actual data you have to start with? That was very unclear.
Excellent idea. We have implemented this in Figure 1.
Labeling session by "group" is very confusing. I think most readers take "group" as the group of subjects, but that's not what you mean at all. You mean some sessions were one way and some were another. (And, as I noted in the public review, you ignore many of the sessions, which I think is not OK.) I think a major rewrite would help a lot. Also, I don't think the group analysis is necessary at all. In the public review, I recommend doing the analyses very differently and more classically.
We have updated the group assignments in a manner that is more intuitive, reflects the delays, and includes all sessions.
The paper is full of arbitrary abbreviations that are completely unnecessary. Every time I came to "ival", I had to translate that into "number of pellets delivered on the immediate lever" and every time I came to dLP, I had to translate that into "delayed lever press". Making the text shorter does not make the text easier to read. In general, I was taught that unless the abbreviation is the common term (such as "DNA" not "deoxyribonucleic acid"), you should never use an abbreviation. While there are some edge cases (ACC probably over "anterior cingulate cortex"), dLP, iLP, dLPs, iLPs, ival, are definitely way over the "don't do that" line.
We completely agree here and apologize for the excessive use of abbreviations. We have removed nearly all of them
The figures were incomplete, poorly labeled, and hard to read. A lot of figures were missing, for example
Basic task structure
Basic behavior on the task
Scatter plot of the measures that you are clustering (lever press choice X number of pellets on the immediate lever, you can use color or multiple panels to indicate the delay to the delayed lever) Figure 3 is just a couple of examples. That isn't convincing at all.
Figure 4 is missing labels. In Figure 4, I don't understand what you are trying to say.
I don't see how the results on page 16 arise from Figure 6. I strongly recommend starting from the actual data and working your way to what it means rather than forcing this into this unreasonable "session group" analysis.
We have completely reworked the Figures for clarity and content.
The statement that "no prior study has explored the cellular correlates of cognitive effort" is ludicrous and insulting. There are dozens of experiments looking at ACC in cognitive effort tasks, in humans, other primates, and rodents. There are many dozens of experiments looking at cellular correlates in intertemporal choice tasks, some with neural manipulations, some with ensemble recordings. There are many dozens of experiments looking at cellular relationships to waiting out a delay.
We agree that our statement was extremely imprecise. We have updated this to say: “Further, a role for theta oscillations in allocating physical effort has been identified. However, the cellular
mechanisms within the ACC that control and deploy types of cognitive effort have not been identified.”
Reviewer #2 (Recommendations For The Authors):
In Figure 2, the panels below E and F are referred to as 'right' - but they are below? I would give them letters.
I would make sure that animal #s, neuron #s, and LFP#s are clearly presented in the results and in each figure legend. This is important to follow the results throughout the manuscript.
Some additional proofreading ('Fronotmedial') might help with clarity.
Based on our updates, this is no longer relevant.
Reviewer #3 (Recommendations For The Authors):
In addition to the suggestions above to address specific issues, it would be useful to report some additional information about aspects of the experiments and analyses:
Specify how spike sorting was performed and what metrics were used to select well isolated single units.
Done.
Provide histology showing the recording locations for each subject.
Histological assessments of electrodes placements are provided in White et al. 2024, but we provide an example placement. This has been added to the text.
Indicate the sequence of recording sessions that occurred for each subject, including for each session what delay duration was used and which dataset the session contributed to, and indicate when the neural probes were advanced between sessions.
We feel that this adds complexity unnecessarily as we make no claims about holding units across sessions for differences in coding in the dorsoventral gradient of ACC.
Indicate the experimental unit when reporting uncertainty measures in figure legends (e.g. mean +/- SEM across sessions).
Done.
-
-
www.trs-80.org www.trs-80.org
-
He looked at me incredulously and said, “Fortran is a compiler. It’s a computer program like any other. Only it happens to take source code as its input, and puts out machine code.” There was a long pause, then I said, “Someone wrote that program?” I was stunned. I don’t know where I thought the compiler came from — Mount Olympus, maybe?
Crenshaw's humility notwithstanding, I'm frequently caught off guard by frequent interactions with people whose conversational posture reveals that they have a similar conception of software like, say, Windows—their comments a manifestation of a seemingly total unwillingness to confront the fact that, no, some observable behavior in software isn't just how computers work, but that someone—a human programmer—sat down and decided to make it work that way—that it isn't just some natural property of computers that someone has coaxed out of one, and that other instances of software creation are not mere parlor tricks. They're procedures. They have to be conceived of and then worked out and (ideally) made airtight against a whole range of conditions.
-
-
www.cartoonshateher.com www.cartoonshateher.com
-
Why do you even want to get married? It’s just a piece of paper. (Often said by people who were married themselves, acting like marriage happened to them by mistake).Four years isn’t that long. You’re so young!Women have so many better options than marriage, why don’t you travel the world instead?At 23, you don’t even know yourself.If you want to get married this badly, you don’t really love your boyfriend, you just love weddings.You should never talk to him about your marriage timeline because he will think you’re crazy and clingy.You shouldn’t care when (nay, if) he proposes at all.If you’re insecure about your relationship that means you need to go to therapy to cure all your negative emotions once and for all.
-
-
socialsci.libretexts.org socialsci.libretexts.org
-
With single parenting and cohabitation (when a couple shares a residence but not a marriage) becoming more acceptable in recent years, people may be less motivated to get married. In a recent survey, 39 percent of respondents answered “yes” when asked whether marriage is becoming obsolete (Pew Research Center 2010). The institution of marriage is likely to continue, but some previous patterns of marriage will become outdated as new patterns emerge. In this context, cohabitation contributes to the phenomenon of people getting married for the first time at a later age than was typical in earlier generations (Glezer 1991). Furthermore, marriage will continue to be delayed as more people place education and career ahead of “settling down.”
The people that are just OK with cohabitation rather than marriage surprise me but then it makes sense when we get to the next Section and they talk about one partner or many. The reason why more people are OK with cohabitation is because it’s less commitment rather than actually proposing a marriage, people want to still feel like they have the option to leave.(so they don’t feel trapped) It’s like they’re afraid of not having freedom or just afraid of the commitment in general.
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the previous reviews.
eLife Assessment:
Glioblastoma is one of the most aggressive cancers without a cure. Glioblastoma cells are known to have high mitochondrial potential. This useful study demonstrates the critical role of the ribosome-associated quality control (RQC) pathway in regulating mitochondrial membrane potential and glioblastoma growth. Some assays are incomplete; further revision will improve the significance of this study.
For clarity, we propose revising the second sentence to: "It is well-established that certain cancer cells, such as glioblastoma cells, exhibit elevated mitochondrial membrane potential."
Reviewer #1 (Public Review):
Summary:
Cai et al have investigated the role of msiCAT-tailed mitochondrial proteins that frequently exist in glioblastoma stem cells. Overexpression of msiCAT-tailed mitochondrial ATP synthase F1 subunit alpha (ATP5) protein increases the mitochondrial membrane potential and blocks mitochondrial permeability transition pore formation/opening. These changes in mitochondrial properties provide resistance to staurosporine (STS)-induced apoptosis in GBM cells. Therefore, msiCAT-tailing can promote cell survival and migration, while genetic and pharmacological inhibition of msiCAT-tailing can prevent the overgrowth of GBM cells.
Strengths:
The CAT-tailing concept has not been explored in cancer settings. Therefore, the present provides new insights for widening the therapeutic avenue.
Your acknowledgment of our study's pioneering elements is greatly appreciated.
Weaknesses:
Although the paper does have strengths in principle, the weaknesses of the paper are that these strengths are not directly demonstrated. The conclusions of this paper are mostly well-supported by data, but some aspects of image acquisition and data analysis need to be clarified and extended.
We are grateful for your acknowledgment of our study’s innovative approach and its possible influence on cancer therapy. We sincerely appreciate your valuable feedback. In response, this updated manuscript presents substantial new findings that reinforce our central argument. Moreover, we have broadened our data analysis and interpretation, as well as refined our methodological descriptions.
Reviewer #2 (Public Review):
This work explores the connection between glioblastoma, mito-RQC, and msiCAT-tailing. They build upon previous work concluding that ATP5alpha is CAT-tailed and explore how CAT-tailing may affect cell physiology and sensitivity to chemotherapy. The authors conclude that when ATP5alpha is CAT-tailed, it either incorporates into the proton pump or aggregates and that these events dysregulate MPTP opening and mitochondrial membrane potential and that this regulates drug sensitivity. This work includes several intriguing and novel observations connecting cell physiology, RQC, and drug sensitivity. This is also the first time this reviewer has seen an investigation of how a CAT tail may specifically affect the function of a protein. However, some of the conclusions in this work are not well supported. This significantly weakens the work but can be addressed through further experiments or by weakening the text.
We appreciate the recognition of our study's novelty. To address your concerns about our conclusions, we have revised the manuscript. This revision includes new data and corrections of identified issues. Our detailed responses to your specific points are outlined below.
Reviewer #1 (Recommendations For The Authors):
(1) In Figure 1B, please replace the high-exposure blots of ATP5 and COX with representative results. The current results are difficult to interpret clearly. Additionally, it would be helpful if the author could explain the nature of the two different bands in NEMF and ANKZF1. Did the authors also examine other RQC factors and mitochondrial ETC proteins? I'm also curious to understand why CAT-tailing is specific to C-I30, ATP5, and COX-V, and why the authors did not show the significance of COX-V.
We appreciate your inquiry regarding the data. Additional attempts were made using new patient-derived samples; however, these results did not improve upon the existing ATP5⍺, (NDUS3)C-I30, and COX4 signals presented in the figure. This is possibly due to the fact that CAT-tail modified mitochondrial proteins represent only a small fraction of the total proteins in these cells. It is acknowledged that the small tails visible above the prominent main bands are not particularly distinct. To address this, the revised version includes updated images to better illustrate the differences. We believe the assertion that GBM/GSCs possess CAT-tailed proteins is substantiated by a combination of subsequent experimental findings. The figure (refer to new Fig. 1B) serves primarily as an introduction. It is important to note that the CAT-tailed ATP5⍺ plays a vital role in modulating mitochondrial potential and glioma phenotypes, a function which has been demonstrated through subsequent experiments.
It is acknowledged that the CAT-tail modification is not exclusive to the ATP5⍺protein. ATP5⍺ was selected as the primary focus of this study due to its prevalence in mitochondria and its specific involvement in cancer development, as noted by Chang YW et al. Future research will explore the possibility of CAT tails on other mitochondrial ETC proteins. Currently, NDUS3 (C-I30), ATP5⍺, and COX4 serve as examples confirming the existence of these modifications. It remains challenging to detect endogenous CAT-tailing, and bulk proteomics is not yet feasible for this purpose. COX4 is considered significant. We hypothesize that CAT-tailed COX4 may function similarly to the previously studied C-I30 (Wu Z, et al), potentially causing substantial mitochondrial proteostasis stress.
Concerning RQC proteins, our blotting analysis of GBM cell lines now includes additional RQC-related factors. The primary, more prominent bands (indicated by arrowheads) are, in our assessment, the intended bands for NEMF and ANKZF1. Subsequent blotting analyses showed only single bands for both ANKZF1 and NEMF, respectively. The additional, larger molecular weight band of NEMF, which was initially considered for property analysis (phosphorylation, ubiquitination, etc.), was not examined further as it did not appear in subsequent experiments (refer to new Fig. S1C).
References:
Chang YW, et al. Spatial and temporal dynamics of ATP synthase from mitochondria toward the cell surface. Communications biology. 2023;6(1).
Wu Z, et al. MISTERMINATE Mechanistically Links Mitochondrial Dysfunction With Proteostasis Failure. Molecular cell. 2019;75(4).
(2) In addition to Figure 1B, it would be interesting to explore CAT-tailed mETC proteins in cancer tissue samples.
This is an excellent point, and we appreciate the question. We conducted staining for ATP5⍺ and key RQC proteins in both tumor and normal mouse tissues. Notably, ATP5⍺ in GBM exhibited a greater tendency to form clustered punctate patterns compared to normal brain tissue, and not all of it co-localized with the mitochondrial marker TOM20 (refer to new Fig. S3C-E). Crucially, we observed a significant increase in NEMF expression within mouse xenograft tumor tissues, alongside a decrease in ANKZF1 expression (refer to new Fig. S1A, B). These findings align with our observations in human samples.
(3) Please knock down ATP5 in the patient's cells and check whether both the upper band and lower band of ATP5 have disappeared or not.
This control was essential and has been executed now. To validate the antibody's specificity, siRNA knockdown was performed. The simultaneous elimination of both upper and lower bands upon siRNA treatment (refer to new Fig. S2A) confirms they represent genuine signals recognized by the antibody.
(4) In Figure 1C and ID, add long exposure to spot aggregation and oligomer. Figure 1D, please add the blots where control and ATP5 are also shown in NHA and SF (similar to SVG and GSC827).
New data are included in the revised manuscript to address the queries. Specifically, the new Fig 1D now displays the full queue as requested, featuring blots for Control, ATP5α, AT3, and AT20. Our analysis reveals that AT20 aggregates exhibit higher expression and accumulation rates in GSC and SF cells.
Fig. 1C has been updated to include experimental groups treated with cycloheximide and sgNEMF. Our results show that sgNEMF effectively inhibits CAT-tailing in GBM cell lines, whereas cycloheximide has no impact. After consulting with the Reporter's original creator and optimizing expression conditions, we observed no significant aggregates with β-globin-non-stop protein, potentially due to the length of endogenous CAT-tail formation (as noted by Inada, 2020, in Cell Reports). Our analysis focused on the ratio of CAT-tailed (red box blots) and non-CAT-tailed proteins (green box blots). Comparing these ratios revealed that both anisomycin treatment and sgNEMF effectively hinder the CAT-tailing process, while cycloheximide has no effect.
(5) In Figure 1E, please double-check the results with the figure legend. ATP5A aggregated should be shown endogenously. The number of aggregates shown in the bar graph is not represented in micrographs. Please replace the images. For Figure 1E, to confirm the ATP5-specific aggregates, it would be better if the authors would show endogenous immunostaining of C-130 and Cox-IV.
Labels in Fig. 1E were corrected to reflect that the bar graph in Fig. 1F indicates the number of cells with aggregates, not the quantity of aggregates per cell. The presence
(6) Figure 3A. Please add representative images in the anisomycin sections. It is difficult to address the difference.
We appreciate your feedback. Upon re-examining the Calcein fluorescence intensity data in Fig. 3A, we believe the images accurately represent the statistical variations presented in Fig. 3B. To address your concerns more effectively, please specify which signals in Fig. 3A you find potentially misleading. We are prepared to revise or substitute those images accordingly.
(7) Figure 3D. If NEMF is overexpressed, is the CAT-tailing of ATP 5 reversed?
Thank you. Your prediction aligns with our findings. We've added data to the revised Fig. S6A, B, which demonstrates that both NEMF overexpression and ANKZF1 knockdown lead to elevated levels of CRC. This increase, however, was not statistically significant in GSC cells. A plausible explanation for this discrepancy is that the MPTP of GSC cells is already closed, thus any additional increase in CAT-tailing activity does not result in further amplification.
(8) Figure 3G. Why on the BN page are AT20 aggregates not the same as shown in Figure 2E?
We appreciate your inquiry regarding the ATP5⍺ blots, specifically those in the original Fig. 3G (left) and 2E (right). Careful observation of the ATP5⍺ band placement in these figures reveals a high degree of similarity. Notably, there are aggregates present at the top, and the diffuse signals extend downwards. Given that this is a gradient polyacrylamide native PAGE, the concentration diminishes towards the top. Consequently, the non-rigid nature of the Blue Native PAGE gel may lead to slight variations in the aggregate signals; however, the overall patterns are very much alike. To mitigate potential misinterpretations, we have rearranged the blot order in the new Fig. 3M.
(9) Figure 4D. The amount of aggregation mediated by AT20 is more compared to AT3. Why are there no such drastic effects observed between AT3 and AT20 in the Tunnel assay?
The previous Figure 4D presents the quantification of cell migration from the experiment depicted in Figure 4C. But this is a good point. TUNEL staining results are directly influenced by mitochondrial membrane potential and the state of mitochondrial permeability transition pores
(MPTP), not by the degree of protein aggregation. Our previous experiments showed comparable effects of AT3 and AT20 on mitochondria (Fig. 2E, 3K), which aligns with the expected similar outcomes on TUNEL staining. As for its biological nature, this could be very complicated. We hope to explore it in future studies.
(10) Figure 5C: The role of NEMF and ANKZF1 can be further clarified by conducting Annexin-PI assays using FACS. The inclusion of these additional data points will provide more robust evidence for CAT-tailing's role in cancer cells.
In response to your suggestion, we have incorporated additional data into the revised version.Using the Annexin-PI kit, we labeled apoptotic cells and detected them using flow cytometry (FACS). Our findings indicate that anisomycin pretreatment, NEMF knockdown (sgNEMF), and ANZKF1 upregulation (oeANKZF1) significantly increase the rate of STS-induced apoptosis compared to the control group (refer to new Fig. S9D-G).
(11) Figure 5F: STS is a known apoptosis inhibitor. Why it is not showing PARP cleavage? Also, cell death analysis would be more pronounced, if it could be shown at a later time point. What is the STS and Anisomycin at 24h or 48h time-point? Since PARP is cleaved, it would also be better if the authors could include caspase blots.
I guess what you meant to say here is "Staurosporine is a protein kinase inhibitor that can induce apoptosis in multiple mammalian cell lines." Our study observed PARP cleavage even in GSCs, which are typically more resistant to staurosporine-induced apoptosis (C-PARP in Fig. S9B). The ratio of C-PARP to total PARP increased. We selected a 180-minute treatment duration because longer treatments with STS + anisomycin led to a late stage of apoptosis and non-specific protein degradation (e.g., at 24 or 48 hours), making PARP comparisons less meaningful. Following your suggestion, we also examined caspase 3/7 activity in GSC cells treated with DMSO, CHX, and anisomycin. We found that anisomycin treatment also activated caspases (Fig. S9A).
(12) In Figure 5, the addition of an explanation, how CAT-tailing can induce cell death, would add more information such as BAX-BCL2 ratio, and cytochrome-c release from the mitochondria.
Thank you for your suggestion. In this study, we state that specific CAT-tails inhibit GSC cell death/apoptosis rather than inducing it. Therefore, we do not expect that examining BAX-BCL2 and mitochondrial cytochrome c release would offer additional insights.
(13) To confirm the STS resistance, it would be better if the author could do the experiments in the STS-resistant cell line and then perform the Anisomycin experiments.
Thank you. We should emphasize that our data primarily originates from GSC cells. These cells already exhibit STS-resistance when compared to the control cells (Fig. S8A-C).
(14) It would be more advantageous if the author could show ATP5 CATailed status under standard chemotherapy conditions in either cell lines or in vivo conditions.
This is an interesting question. It's worth exploring this question; however, GSC cells exhibit strong resistance to standard chemotherapy treatments like temozolomide (TMZ).
Additionally, we couldn't detect changes in CAT-tailed ATP5⍺ and thus did not include that data.
(15) In vivo (cancer mouse model or cancer fly model) data will add more weight to the story.
We appreciate your intriguing question. An effective approach would be to test the RQC pathway's function using the Drosophila Notch overexpression-induced brain tumor model. However, Khaket et al. have conducted similar studies, stating, "The RNAi of Clbn, VCP, and Listerin (Ltn), homologs of key components of the yeast RQC machinery, all attenuated NSC over-proliferation induced by Notch OE (Figs. 5A and S5A–D, G)." This data supports our theory, and we have incorporated it into the Discussion. While the mouse model more closely resembles the clinical setting, it is not covered by our current IACUC proposal. We intend to verify this hypothesis in a future study.
Reference:
Khaket TP, Rimal S, Wang X, Bhurtel S, Wu YC, Lu B. Ribosome stalling during c-myc translation presents actionable cancer cell vulnerability. PNAS Nexus. 2024 Aug 13;3(8):pgae321.
Reviewer #2 (Recommendations For The Authors):
Figure 1B, C: To demonstrate that Globin, ATP5alpha, and C-130 are CAT-tailed, it is necessary to show that the high mobility band disappears after NEMF deletion or mutagenesis of the NFACT domain of NEMF. This can be done in a cell line. The anisomycin experiment is not convincing because the intensity of the bands drops and because no control is done to show that the effects are not due to translation inhibition (e.g. cycloheximide, which inhibits translation but not CAT tailing). Establishing ATP5alpha as a bonafide RQC substrate and CAT-tailed protein is critical to the relevance of the rest of the paper.
Thank you for suggesting this crucial control experiment. To confirm the observed signal is indeed a bona fide CAT-tail, it's essential to demonstrate that NEMF is necessary for the CAT-tailing process. We have incorporated data from NEMF knockdown (sgNEMF) and cycloheximide treatment into the revised manuscript. Our findings show that both sgNEMF and anisomycin treatment effectively inhibit the formation of CAT-tailing signals on the reporter protein (Fig. 1C). Similarly, NEMF knockdown in a GSC cell line also effectively eliminated CAT-tails on overexpressed ATP5⍺ (Fig. S2B).
In general, the text should be weakened to reflect that conclusions were largely gleaned from artificial CAT tails made of AT repeats rather than endogenously CAT-tailed ATP5alpha. CAT tails could have other sequences or be made of pure alanine, as has been suggested by some studies.
Thank you for your reminder. We have reviewed the recent studies by Khan et al. and Chang et al., and we found their analysis of CAT tail components to be highly insightful. We concur with your suggestion regarding the design of the CAT tail sequence. We aimed to design a tail that maintained stability and resisted rapid degradation, regardless of its length. In the revised version, we clarify that our conclusions are based on artificial CAT tails, specifically those composed of AT repeat sequences (p. 9). We acknowledge that the presence of other sequence components may lead to different outcomes (p. 19).
Reference:
Khan D, Vinayak AA, Sitron CS, Brandman O. Mechanochemical forces regulate the composition and fate of stalled nascent chains. bioRxiv [Preprint]. 2024 Oct 14:2024.08.02.606406. Chang WD, Yoon MJ, Yeo KH, Choe YJ. Threonine-rich carboxyl-terminal extension drives aggregation of stalled polypeptides. Mol Cell. 2024 Nov 21;84(22):4334-4349.e7.
Throughout the work (e.g. 3B, C), anisomycin effects should be compared to those with cycloheximide to observe if the effects are specific to a CAT tail inhibitor rather than a translation inhibitor.
We agree that including cycloheximide control experiments is crucial. The revised version now incorporates new data, as depicted in Fig. S5A, B, illustrating alterations in the on/off state of MPTP following cycloheximide treatment. Furthermore, Fig. S6A, B present changes in Calcium Retention Capacity (CRC) under cycloheximide treatment. The consistency of results across these experiments, despite cycloheximide treatment, suggests that anisomycin's role is specifically as a CAT tail inhibitor, rather than a translation inhibitor.
Line 110, it is unclear what "short-tailed ATP5" is. Do you mean ATP5alpha-AT3? If so this needs to be introduced properly. Line 132: should say "may indicate accumulation of CAT-tailed protein" rather than "imply".
We acknowledge your points. We have clarified that the "short-tailed ATP5α" refers to ATP5α-AT3 and incorporated the requested changes into the revised manuscript.
Figure 1C: how big are those potential CAT-tails (need to be verified as mentioned earlier)?They look gigantic. Include a ladder.
In the revised Fig. 1D, molecular weight markers have been included to denote signal sizes. The aggregates in the previous Fig. 1C, also present in the control plasmid, are likely a result of signal overexposure. The CAT-tailed protein is observed just above the intended band in these blots. These aggregates have been re-presented in the updated figures, and their signal intensities quantified.
Line 170: "indicating that GBM cells have more capability to deal with protein aggregation". This logic is unclear. Please explain.
We appreciate your question and have thoroughly re-evaluated our conclusion. We offer several potential explanations for the data presented in Fig. 1D: (1) ATP5α-AT20 may demonstrate superior stability. (2) GSC (GBM) cells might lack adequate mechanisms to monitor protein accumulation. (3) GSC (GBM) cells could possess an increased adaptive capacity to the toxicity arising from protein accumulation. This discussion has been incorporated into the revised manuscript (lines 166-169).
Line 177: how do you know the endogenous ATP5alpha forms aggregates due to CAT-tailing? Need to measure in a NEMF hypomorph.
We understand your concern and have addressed it. Revised Fig. 3G, H demonstrates that a reduction in NEMF levels, achieved through sgNEMF in GSC cells, significantly diminishes ATP5α aggregation. This, in conjunction with the Anisomycin treatment data presented in revised Fig. 3E, F, confirms the substantial impact of the CAT-tailing process on this aggregation.
Line 218: really need a cycloheximide or NEMF hypomorph control to show this specific to CAT-tailing.
We have revised the manuscript to include data from sgNEMF and cycloheximide treatments, specifically Fig. 3G, H, and Fig. S5C, D, as detailed in our response above.
Lines 249,266, Figure 5A: The mentioned experiments would benefit from controls including an extension of ATP5alpha that was not alanine and threonine, perhaps a gly-ser linker, as well as an NEMF hypomorph.
We sincerely appreciate your insightful comments. In response, the revised manuscript now incorporates control data for ATP5α featuring a poly-glycine-serine (GS) tail. This data is specifically presented in Figs. S2E-G, S4E, S7A, D, E, and S8F, G. Our experimental findings consistently demonstrate that the overexpression of ATP5α, when modified with GS tails, had no discernible impact on protein aggregation, mitochondrial membrane potential, GSC cell mobility, or any other indicators assessed in our study.
Figure S5A should be part of the main figures and not in the supplement.
This has been moved to the main figure (Fig. 5C).
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public review):
From my reading, this study aimed to achieve two things:
(1) A neurally-informed account of how Pieron's and Fechner's laws can apply in concert at distinct processing levels.
(2) A comprehensive map in time and space of all neural events intervening between stimulus and response in an immediately-reported perceptual decision.
I believe that the authors achieved the first point, mainly owing to a clever contrast comparison paradigm, but with good help also from a new topographic parsing algorithm they created. With this, they found that the time intervening between an early initial sensory evoked potential and an "N2" type process associated with launching the decision process varies inversely with contrast according to Pieron's law. Meanwhile, the interval from that second event up to a neural event peaking just before response increases with contrast, fitting Fechner's law, and a very nice finding is that a diffusion model whose drift rates are scaled by Fechner's law, fit to RT, predicts the observed proportion of correct responses very well. These are all strengths of the study.
We thank the reviewer for their comments that added context to the events we detected in relation to previous findings. We also believe that the change in the HMP algorithm suggested by the reviewer improved the precision of our analyses and the manuscript. We respond to the reviewer’s specific comments below.
(1) The second, generally stated aim above is, in the opinion of this reviewer, unconvincing and ill-defined. Presumably, the full sequence of neural events is massively task-dependent, and surely it is more in number than just three. Even the sensory evoked potential typically observed for average ERPs, even for passive viewing, would include a series of 3 or more components - C1, P1, N1, etc. So are some events being missed? Perhaps the authors are identifying key events that impressively demarcate Pieron- and Fechner-adherent sections of the RT, but they might want to temper the claim that they are finding ALL events. In addition, the propensity for topographic parsing algorithms to potentially lump together distinct processes that partially co-evolve should be acknowledged.
We agree with the reviewer that the topographical solutions found by HMP will be dependent on the task and the quality and type of data. We address this point in the last section of the discussion (see also response to R3.5). We would also like to add that the events detected by HMP are, by construction, those that contribute to the RT and not necessarily all ERPs elicited by a stimulus.
In addition to the new last section of the discussion we also make these points clear in the revised manuscript at the discussion start:
“By modeling the recorded single-trial EEG signal between stimulus onset and response as a sequence of multivariate events with varying by-trial peak times, we aimed to detect recurrent events that contribute to the duration of the reaction time in the present perceptual decision-making task”.
Regarding the typical visual ERPs, in response to this comment but also comments R1.2, R1.3 and R2.1, we aimed for a more precise description of the topographies and thus reduced the width of the HMP expected events to 25ms. This ensures that we do not miss events shorter than the initial expectations of 50ms (see Appendix B of Weindel et al., 2024 and also response to R1.3). This new estimation provides evidence for at least two of the visual ERPs that, based on their timings and topographies (in relation with the spatial frequency of the stimulus), we interpret as the N40 and the P100 (see response to R1.5 for the justification of this categorization). We provide a description and justification of the interpretations in the result section “Five trial-recurrent sequential events occur in the EEG during decisions” and the discussion section “Visual encoding time”.
(2) To take a salient example, the last neural event seems to blend the centroparietal positivity with a more frontal midline negativity, some of which would capture the CNV and some motor-execution related components that are more tightly time-locked to, of course, the response. If the authors plotted the traditional single-electrode ERP at the frontal focus and centroparietal focus separately, they are likely to see very different dynamics and contrast- and SAT-dependency. What does this mean for the validity of the multivariate method? If two or more components are being lumped into one neural event, wouldn't it mean that properties of one (e.g., frontal burstiness at response) are being misattributed to the other (centroparietal signal that also peaks but less sharply at response)?
Using the new HMP parameterization described above we show that the reviewer's intuition was correct. Using an expected pattern duration of 25ms the last event in the original manuscript splits in two events. The before-last event, now referred to the lateralized readiness potential (LRP) presents a strong lateralization (Figure 3) with an increased negativity over the motor cortex contralateral to the right hand. The effect of contrast is mostly on the last event that we interpret as the CPP (Figure 5). Despite the improved precision of the topographies of the identified events, it is however to be noted that some components will overlap. If the LRP is generated when a certain amount of evidence is accumulated (e.g. that the CPP crosses a certain value) then a time-based topography will necessarily include that CPP activity in addition to the lateralized potential. We discuss this in the section “Motor execution” of the discussion:
“Adding the abrupt onset of this potential, we believe that this event is the start of motor execution, engaged after a certain amount of evidence. The evidence for this interpretation is manifest in the fact that the event's topography shares some activity with the CPP event that follows, an expected result if the LRP is triggered at a certain amount of evidence, indexed by the CPP”.
(3) Also related to the method, why must the neural events all be 50 ms wide, and what happens if that is changed? Is it realistic that these neural events would be the same duration on every trial, even if their duration was a free parameter? This might be reasonable for sensory and motor components, but unlikely for cognitive.
The HMP method is sensitive to the event's duration as shown in the manuscript about the method (Appendix B of Weindel et al., 2024). Nevertheless as long as the topography in the real data is longer than the expected one it shouldn't be missed (i.e. same goes for by-trial variations in the event width). For this reason we halved the expected event width of 50ms (introduced by the original HsMM-MVPA paper by Anderson and colleagues) in the revision. This new estimation with 25ms thus is much less likely to miss events as evidenced by the new visual and motor events. In the revised manuscript this is addressed at the start of the Results section:
“Contrary to previous applications (Anderson et al.,2016; Berberyan et al., 2021; Zhang et al., 2018; Krause et al., 2024) we assumed that the multivariate pattern was represented by a 25ms half-sine as our previous research showed that a shorter expected pattern width increases the likelihood of detecting cognitive events (see Appendix B of Weindel et al., 2024)”.
Regarding the event width as a free parameter this is both technically and statistically difficult to implement as the amount of computing capacity, flexibility and trade-offs among the HMP parameters would, given the current implementation, render the model unfit for most computers and statistically unidentifiable.
(4) In general, I wonder about the analytic advantage of the parsing method - the paradigm itself is so well-designed that the story may be clear from standard average event-related potential analysis, and this might sidestep the doubts around whether the algorithm is correctly parsing all neural events.
Average ERP analysis suffers from an impossibility to differentiate between an effect of an experimental factor on the amplitude vs. on the timing of the underlying components (Luck, 2005). Furthermore the overlap of components across trials bluries the distinction between them. For both reasons we would not be able to reach the same level of certainty and precision using ERP analyses. Furthermore the relatively low number of trials per experimental cell (contrast level X SAT X participant = 6 trials) makes the analyses hard to perform on ERP which typically require more trials per modality. From the reviewer’s comment we understand that this point was not clear. We therefore discuss this in the revision, Section “Functional interpretation of the events” of the results:
“Nevertheless identifying neural dynamics on these ERPs centered on stimulus is complicated by the time variation of the underlying single-trial events (see probabilities displayed in Figure 3 for an illustration and Burle et al., 2008, for a discussion). The likely impact of contrast on both amplitude and time on the underlying single-trial event does not allow one to interpret the average ERP traces as showing an effect in one or the other dimension without strong assumptions (Luck, 2005)”.
(5) In particular, would the authors consider plotting CPP waveforms in the traditional way, across contrast levels? The elegant design is such that the C1 component (which has similar topography) will show up negative and early, giving way to the CPP, and these two components will show opposite amplitude variations (not just temporal intervals as is this paper's main focus), because the brighter the two gratings, the stronger the aggregate early sensory response but the weaker the decision evidence due to Fechner. I believe this would provide a simple, helpful corroborating analysis to back up the main functional interpretation in the paper.
We agree with the suggestion and have introduced the representation on top of Figure 5 for sets of three electrodes in the occipital, posterior and frontal regions. The new panels clearly show an inversion of the contrast effect dependent on the time and locus of the electrodes. We discuss this in Section “Functional interpretation of the events” of the results:
“This representation shows that there is an inversion of the contrast effect with higher contrasts having a higher amplitude on the electrodes associated with visual potentials in the first couple of deciseconds (left panel of Figure 5A) while parietal and frontal electrodes shows a higher amplitude for lower contrasts in later portions of the ERPs (middle and right panel of Figure 5A)”.
To us, this crucially shows that we cannot achieve the same decomposition using traditional ERP analyses. In these plots it appears that while, as described by the reviewer, there is an inversion, the timing and amplitude of the changes due to contrast can hardly be interpreted.
(6) The first component is picking up on the C1 component (which is negative for these stimulus locations), not a "P100". Please consult any visual evoked potential study (e.g., Luck, Hillyard, etc). It is unexpected that this does not vary in latency with contrast - see, for example. Gebodh et al (2017, Brain Topography) - and there is little discussion of this. Could it be that nonlinear trends were not correctly tested for?
We disagree with the reviewer on the interpretation of the ERP. The timing of the detected component is later than the one usually associated with a C1. Furthermore the central display does not create optimal conditions to detect a C1
We do agree that the topography raises the confusion but we believe that this is due to the spatial frequency of the stimulus that generates a high posterior positivity (see references in the following extract). The new HMP solution also now happens to show an effect of contrast on the P100 latencies, we believe this is due to the increased precision in the time location of the component. We discuss this in the “Visual encoding time” section of the discussion:
“The following event, the P100, is expressed around 70ms after the N40, its topography is congruent with reports for stimuli with low spatial frequencies as used in the current study (Kenemans et al., 2002, 2000; Proverbio et al., 1996). The timing of this P100 component is changed by the contrast of the stimulus in the direction expected by the Piéron law (Figure 4A)”.
(7) There is very little analysis or discussion of the second stage linked to attention orientation - what would the role of attention orientation be in this task? Is it spatial attention directed to the higher contrast grating (and if so, should it lateralise accordingly?), or is it more of an alerting function the authors have in mind here?
We agree that we were not specific enough on the interpretation of this attention stage. We now discuss our hypothesis in the section “Attention orientation” of the discussion:
“We do however observe an asymmetry in the topographical map Figure 3. This asymmetry might point to an attentional bias with participants (or at least some participants) allocating attention to one side over the other in the same way as the N2pc component (Luck and Hillyard, 1994, Luck et al., 1997). Based on this collection of observations, we conclude that this third event represents an attention orientation process. In line with the finding of Philiastides et al. (2006), this attention orientation event might also relate to the allocation of resources. Other designs varying the expected cognitive load or spatial attention could help in further interpreting the functional role of this third event”.
We would like to add that it is unlikely that the asymmetry we mention in the discussion cannot stem from the redirection towards higher contrast as the experimental design balanced the side of presentation. We therefore believe that this is a behavioral bias rather than a bias toward the highest contrast stimulus as suggested by the reviewer. We hope that, while more could be tested and discussed, this discussion is sufficient given the current manuscript's goal.
Reviewer #2 (Public review):
Summary:
The authors decomposed response times into component processes and manipulated the duration of these processes in opposing directions by varying contrast, and overall by manipulating speed-accuracy tradeoffs. They identify different processes and their durations by identifying neural states in time and validate their functional significance by showing that their properties vary selectively as expected with the predicted effects of the contrast manipulation. They identify 3 processes: stimulus encoding, attention orienting, and decision. These map onto classical event-related potentials. The decision-making component matched the CPP, and its properties varied with contrast and predicted decision-accuracy, while also exhibiting a burst not characteristic of evidence accumulation.
Strengths:
The design of the experiment is remarkable and offers crucial insights. The analysis techniques are beyond state-of-the-art, and the analyses are well motivated and offer clear insights.
Weaknesses:
It is not clear to me that the results confirm that there are only 3 processes, since e.g., motor preparation and execution were not captured. While the authors discuss this, this is a clear weakness of the approach, as other components may also have been missed. It is also unclear to what extent topographies map onto processes, since, e.g., different combinations of sources can lead to the same scalp topography.
We thank the reviewer for their kind words and for the attention they brought on the question of the missing motor preparation event. In light of this comment (and also R1.1, R3.3) the revised manuscript uses a finer grained approach for the multivariate event detection. This preciser estimation comes from the use of a shorter expected pattern in which the initial expectation of a 50ms half-sine was halved, therefore ensuring that we do not miss events shorter than the initial expectations (see Appendix B of Weindel et al., 2024 and also response to R1.3). In the new solution the motor component that the reviewer expected is found as evidenced by the topography of the event, its lateralization and a time-to-response congruent with a response execution event. This is now described in the section “Motor execution” of the revised manuscript:
“The before last event, identified as the LRP, shows a strong hemispheric asymmetry congruent with a right hand response. The peak of this event is approximately 100 ms before the response which is congruent with reports that the LRP peaks at the onset of electromyographical activity in the effector muscle (Burle et al., 2004), typically happening 100ms before the response in such decision-making tasks (Weindel et al., 2021). Furthermore, while its peak time is dependent on contrast, its expression in the EEG is less clearly related to the contrast manipulation than the following CPP event”.
Reviewer #3 (Public review):
Summary:
In this manuscript, the authors examine the processing stages involved in perceptual decision-making using a new approach to analysing EEG data, combined with a critical stimulus manipulation. This new EEG analysis method enables single-trial estimates of the timing and amplitude of transient changes in EEG time-series, recurrent across trials in a behavioural task. The authors find evidence for three events between stimulus onset and the response in a two-spatial-interval visual discrimination task. By analysing the timing and amplitude of these events in relation to behaviour and the stimulus manipulation, the authors interpret these events as related to separable processing stages for stimulus encoding, attention orientation, and decision (deliberation). This is largely consistent with previous findings from both event-related potentials (across trials) and single-trial estimates using decoding techniques and neural network approaches.
Strengths:
This work is not only important for the conceptual advance, but also in promoting this new analysis technique, which will likely prove useful in future research. For the broader picture, this work is an excellent example of the utility of neural measures for mental chronometry.
We appreciate the very positive review and thank the reviewer for pointing out important weaknesses in our original manuscript and also providing resources to address them in the recommendations to authors. Below we comment on each identified weakness and how we addressed them.
Weaknesses:
(1) The manuscript would benefit from some conceptual clarifications, which are important for readers to understand this manuscript as a stand-alone work. This includes clearer definitions of Piéron's and Fechner's laws, and a fuller description of the EEG analysis technique.
We agree that the description of both laws were insufficient, we therefore added the following text in the last paragraph of the introduction:
“Piéron’s law predicts that the time to perceive the two stimuli (and thus the choice situation) should follow a negative power law with the stimulus intensity (Figure 1, green curve). In contradistinction, Fechner’s law states that the perceived difference between the two patches follows the logarithm of the absolute contrast of the two patches (Figure 1, yellow curve). As the task of our participants is to judge the contrast difference, Piéron’s law should predict the time at which the comparison starts (i.e. the stimuli become perceptible), while Fechner’s law should implement the comparison, and thus decision, difficulty”.
Regarding the EEG analysis technique we added a few elements at the start of the result:
“The hidden multivariate pattern model (HMP) implemented assumed that a task-related multivariate pattern event is represented by a half-sine whose timing varies from trial to trial based on a gamma distribution with a shape parameter of 2 and a scale, controlling the average latency of the event, free-to-vary per event (Weindel et al., 2024)”.
We also made the technique clearer at the start of the discussion:
“By modeling the recorded single-trial EEG signal between stimulus onset and response as a sequence of multivariate events with varying by-trial peak times, we aimed to detect recurrent events that contribute to the duration of the reaction time in the present perceptual decision-making task. In addition to the number of events, using this hidden multivariate pattern approach (Weindel et al., 2024) we estimated the trial-by-trial probability of each event’s peak, therefore accessing at which time sample each event was the most likely to occur”.
Additionally, we added a proper description in the method section (see the new first paragraph of the “Hidden multivariate pattern” subsection).
(2) The manuscript, broadly, but the introduction especially, may be improved by clearly delineating the multiple aims of this project: examining the processes for decision-making, obtaining single-trial estimates of meaningful EEG-events, and whether central parietal positivity reflects ramping activity or steps averaged across trials.
For the sake of clarity we removed the question of the ramping activity vs steps in the introduction and focused on the processes in decision-making and their single-trial measurement as this is the main topic of the paper. Furthermore the references provided by the reviewer allowed us to write a more comprehensive review of previous studies and how the current study is in line with those. These changes are mainly manifested in these new sentences:
“As an example Philiastides et al. (2006) used a classifier on the EEG activity of several conditions to show that the strength of an early EEG component was proportional to the strength of the stimulus while a later component was related to decision difficulty and behavioral performance (see also Salvador et al., 2022; Philiastides and Sajda, 2006). Furthermore the authors interpreted that a third EEG component was indicative of the resource allocated to the upcoming decision given the perceived decision difficulty. In their study, they showed that it is possible to use single-trial information to separate cognitive processes within decision-making. Nevertheless, their method requires a decoding approach, which requires separate classifiers for each component of interest and restrains the detection of the components to those with decodable discriminating features (e.g. stimuli with strong neural generators such as face stimuli, see Philiastides et al., 2006)”.
(3) A fuller discussion of the limitations of the work, in particular, the absence of motor contributions to reaction time, would also be appreciated.
As laid out in responses to comments R1.1 and R2 the new estimates now include evidence for a motor preparation component. We discuss this in the new “motor execution” paragraph in the discussion section. Additionally we discuss the limitation of the study and the method in the two last paragraphs of the discussion (in the new Section “Generalization and limitation”).
(4) At times, the novelty of the work is perhaps overstated. Rather, readers may appreciate a more comprehensive discussion of the distinctions between the current work and previous techniques to gauge single-trial estimates of decision-related activity, as well as previous findings concerning distinct processing stages in decision-making. Moreover, a discussion of how the events described in this study might generalise to different decision-making tasks in different contexts (for example, in auditory perception, or even value-based decision-making) would also be appreciated.
We agree that the original text could be read as overstating. In addition to the changes linked to R3.2 we also now discuss the link with the previous studies in the before-last paragraph of the discussion before the conclusion in the new “Generalization and limitations” section:
“The present study showed what cognitive processes are contributing to the reaction time and estimated single-trial times of these processes for this specific perceptual decision-making task. The identified processes and topographies ought to be dependent on the task and even the stimuli (e.g. sensory events will change with the sensory modality). More complex designs might generate a higher number of cognitive processes (e.g. memory retrieval from a cue, Anderson et al., 2016) and so could more natural stimuli which might trigger other processes in the EEG (e.g. appraisal vs. choice as shown by Frömer et al., 2024). Nevertheless, the observation of early sensory vs. late decision EEG components is likely to generalize across many stimuli and tasks as it has been observed in other designs and methods (Philiastides et al., 2006; Salvador et al., 2022). To these studies we add that we can evaluate the trial-level contribution, as already done for specific processes (e.g. Si et al., 2020; Sturm et al., 2016), for the collection of events detected in the current study”.
Reviewing Editor Comments:
As you will see, all three reviewers agree that the paper makes a valuable contribution and has many strengths. You will also see that they have provided a range of constructive comments highlighting potential issues with the interpretation of the outcomes of your signal decomposition method. In particular, all three reviewers point out that your results do not identify separate motor preparation signals, which we know must be operating on this type of task. The reviewers suggest further discussion of this issue and the potential limitations of your analysis approach, as well as suggesting some additional analyses that could be run to explore this further. While making these changes would undoubtedly enhance the paper and the final public reviews, I should note that my sense is that they are unlikely to change the reviewers' ratings of the significance of the findings and the strength of evidence in the final eLife assessment
Reviewer #1 (Recommendations for the authors):
(1) Abstract: "choice onset" is ill-defined and not the label most would give the start of the RT interval. Do you mean stimulus onset?
We replaced with "choice onset" with "stimulus onset" in the abstract
(2) Similarly "choice elements" in the introduction seem to refer to sensory attributes/objects being decided about?
We replaced "choice-elements" with "choice-relevant features of the stimuli"
(3) "how the RT emerges from these putative components" - it would be helpful to specify more what level of answer you're looking for, as one could simply answer "when they're done."
We replaced with "how the variability in RTs emerges from these putative components"
(4) Line 61-62: I'm not sure this is a fully correct characterisation of Frömer et al. It was not similar in invoking a step function - it did not invoke any particular mechanism or function, and in that respect does not compare well to Latimer et al. Also, I believe it was the overlap of stimulus-locked components, not response-locked, that they argued could falsely generate accumulator-like buildup in the response-locked ERP.
We indeed wrongly described Frömer et al. The sentence is now "In human EEG data, the classical observation of a slowly evolving centro-parietal positivity, scaling with evidence accumulation, was suggested to result from the overlap of time-varying stimulus-related activity in the response-locked event related potential"
(5) Line 78: Should this be single-trial *latency*?
This referred to location in time but we agree that the term is confusing and thus replaced it with latencies.
(6) The caption of Figure 1 should state what is meant by the y-axis "time"
We added the sentence "The y-axis refers the time predicted by each law given a contrast value (x-axis) and the chosen set of parameters." in the caption of Figure 1
(7) Line 107: Is this the correct description of Fechner's law? If the perceived difference follows the log of the physical difference, then a constant physical difference should mean a constant perceived difference. Perhaps a typo here.
This was indeed a typo we replaced the corresponding part of the sentence with "the perceived difference between the two patches follows the logarithm of the absolute contrast of the two patches"
(8) Line 128: By scale, do you mean magnitude/amplitude?
No, this refers to the parameter of a gamma distribution. To clarify we edited the sentence: "based on a gamma distribution with a shape parameter of 2 and a scale parameter, controlling the average latency of the event, free-to-vary per event"
(9) The caption of Figure 3 is insufficient to make sense of the top panel. What does the inter-event interval mean, and why is it important to show? What is the "response" event?
We agree that the top panel was insufficiently described. To keep the length of the paper short and because of the relatively low amount of information provided by these panels we replaced them for a figure only showing the average topographies as well as the asymmetry tests for each event.
(10) Figure 4: caption should say what the top vs bottom row represents (presumably, accuracy vs speed emphasis?), and what the individual dots represent, given the caption says these are "trial and participant averaged". A legend should be provided for the rightmost panels.
We agree and therefore edited Figure 4. The beginning of the caption mentioned by the reviewer now reads: “A) The panels represent the average duration between events for each contrast level, averaged across participants and trials (stimulus and response respectively as first and last events) for accuracy (top) and speed instructions (bottom).”. Additionally we added legends for the SAT instructions and the model fits.
(11) Line 189: argued for a decision-making role of what?
Stafford and Gurney (2004) proposed that Pieron’s law could reflect a non-linear transformation from sensory input to action outcomes, which they argued reflected a response mechanism. We (Van Maanen et al., 2012) specified this result by showing that a Bayesian Observer Model in which evidence for two alternative options was accumulated following Bayes Rule indeed predicted a power relation between the difference in sensory input of the two alternatives, and mean RT. However, the current data suggest that such an explanation cannot be the full story, as also noted by R3. To clarify this point we replaced the comment by the following sentence:
“Note that this observation is not necessarily incongruent with theoretical work that argued that Piéron’s law could also be a result of a response selection mechanism (Stafford and Gurney, 2004; Van Maanen et al., 2012; Palmer et al., 2005). It could be that differences in stimulus intensity between the two options also contribute to a Piéron-like relationship in the later intervals, that is convoluted with Fechner’s law (see Donkin and Van Maanen, 2014 for a similar argument). Unfortunately, our data do not allow us to discriminate between a pure logarithmic growth function and one that is mediated by a decreasing power function”.
(12) Table 2: There is an SAT effect even on the first interval, which is quite remarkable and could be discussed more - does this mean that the C1 component occurs earlier under speed pressure? This would be the first such finding.
The original event we qualified as a P100 was sensitive to SAT but the earliest event is now the N40 and isn’t statistically sensitive to speed pressure in this data. We believe that the fact that the P100 is still sensitive to SAT is not a surprise and therefore do not outline it.
(13) Line 221: "decrease of activation when contrast (and thus difficulty) increases" - is this shown somewhere in the paper?
The whole section for this analysis was rewritten (see comment below)
(14) I find the analysis of Figure 5 interesting, but the interpretation odd. What is found is that the peak of the decision signal aligns with the response, consistent with previous work, but the authors choose to interpret this as the decision signal "occurring as a short-lived burst." Where is the quantitative analysis of its duration across trials? It can at least be visually appraised in the surface plot, and this shows that the signal has a stimulus-locked onset and, apart from the slowest RTs, remains present and for the most part building, until response. What about this is burst-like? A peak is not a burst.
This was the residue of a previous version of the paper where an analysis reported that no evidence accumulation trace was found. But after proper simulations this analysis turned out to be false because of a poor statistical test. Thus we removed this paragraph in the revised manuscript and Figure 5 has now been extended to include surface plots for all the events.
Reviewer #2 (Recommendations for the authors):
Overall, I really enjoyed reading this paper. However, in some places the approach is a bit opaque or the results are difficult to follow. As I read the paper, I noted:
Did you do a simple DDM, or did you do a collapsing bound for speed?
The fitted DDM was an adaptation of the proportional rate diffusion model. We make this clearer at the end of the introduction: "Given that Fechner’s law is expected to capture decision difficulty we connected this law to the classical diffusion decision models by replacing the rate of accumulation with Fechner’s law in the proportional rate diffusion model of Palmer et al.(2005).”
It is confusing that the order of intervals in the text doesn't match the order in the table. It might be better to say what events the interval is between rather than assuming that the reader reconstructs.
We agree and adapted the order in both the text and the table. The table is now also more explicit (e.g. RT instead of S-R)
Otherwise, I do wonder to what extent the method is able to differentiate processes that yield similar scalp topographies and find it a bit concerning that no motor component was identified.
We believe that the new version with the LRP/CPP is a demonstration that the method can handle similar topographies. The method can handle events with close topographies as long as they are separate in time, however if they are not sequential to one another the method cannot capture both events. We now discuss this, in relation with the C1/P100 overlap, in the discussion section “Visual encoding time”:
“Nevertheless this event, seemingly overlapping with the P100 even at the trial level (Figure 5C), cannot be recovered by the method we applied. The fact that the P100 was recovered instead of the C1 could indicate that only the timing of the P100 contributes to the RT (see Section 3 of Weindel et al., 2024)”.
And we more generally address the question of overlap in the new section “Generalization and limitation”.
Reviewer #3 (Recommendations for the authors):
Major Comments:
(1) If we agree on one thing, it is that motor processes contribute to response time. Line 364: "In the case of decision-making, these discrete neural events are visual encoding, attention-orientation, and decision commitment, and their latency make up the reaction time." Does the third event, "decision commitment", capture both central parietal positivity (decision deliberation) and motor components? If so, how can the authors attribute the effects to decision deliberation as opposed to motor preparation?
Thanks to the suggestions also in the public part. This main problem is now addressed as we do capture both a motor component and a decision commitment.
Line 351 suggests that the third event may contain two components.
This was indeed our initial, badly written, hypothesis. Nevertheless the new solution again addresses this problem.
The time series in Figure 6 shows an additional peak that is not evident in the simulated ramp of Appendix 1.
This was probably due to the overlap of both the CPP and the LRP. It is now much clearer that the CPP looks mostly like a ramp while the LRP looks much more like a burst-like/peaked activity. We make this clear in the “Decision event” paragraph of the discussion section:
“Regarding the build-up of this component, the CPP is seen as originating from single-trial ramping EEG activities but other work (Latimer et al., 2015; Zoltowski et al., 2019) have found support for a discrete event at the trial-level. The ERPs on the trial-by-trial centered event in Figure 5 show support for both accounts. As outlined above, the LRP is indeed a short burst-like activity but the build-up of the CPP between high vs low contrast diverges much earlier than its peak”.
Previous analyses (Weindel et al., 2024) found motor-related activity from central parietal topographies close to the response by comparing the difference in single-trial events on left- vs right-hand response trials. The authors suggest at line 315 that the use of only the right hand for responding prevented them from identifying a motor event.
The use of only the right hand should have made the event more identifiable because the topography would be consistent across trials (rather than inverting on left vs right hand response trials).
The reviewer is correct, in the original manuscript we didn’t test for lateralization, but the comment of the reviewer gave us the idea to explicitly test for the asymmetry (Figure 3). This test now clearly shows what would be expected for a motor event with a strong negativity over the left motor cortex.
The authors state on line 422 that the EEG data were truncated at the time of the response.
Could this have prevented the authors from identifying a motor event that might overlap with the timing of the response?
We thank the reviewer for this suggestion. This would have been a possibility but the problem is that adding samples after the response also adds the post-response processes (error monitoring, button release, stimulus disappearance, etc.). While increasing the samples after the response is definitely something that we need to inspect, we think that the separation we achieved in this revision doesn’t call for this supplementary analysis.
The largest effects of contrast on the third event amplitude appear around the peak as opposed to the ramp. If the peak is caused by the motor component, how does this affect the conclusions that this third event shows a decision-deliberation parietal processes as opposed to a motor process (a number of studies suggest a causal role for motor processes in decision-making e.g. Purcell et al., 2010 Psych Rev; Jun et al., 2021 Nat Neuro; Donner et al., 2009 Curr Bio).
This result now changed and it does look like the peak capturing most of the effect is no longer true. We do however think that there might be some link to theories of motor-related accumulation. We therefore added this to the discussion in the Motor execution section:
“Based on all these observations, it is therefore very likely that this LRP event signs the first passage of a two-step decision process as suggested by recent decision-making models (Servant et al., 2021; Verdonck et al., 2021; Balsdon et al., 2023)”.
I would suggest further investigation into the motor component (perhaps by extending the time window of analysed EEG to a few hundred ms after the response) and at least some discussion of the potential contribution of motor processes, in relation to the previous literature.
We believe that the absence of a motor component is sufficiently addressed in the revised manuscript and in the responses to the other comments.
(2) What do we learn from this work? Readers would appreciate more attention to previous findings and a clearer outline of how this work differs. Two points stand out, outlined below. I believe the authors can address these potential complaints in the introduction and discussion, and perhaps provide some clarification in the presentation of the results.
In the introduction, the authors state that "... to date, no study has been able to provide single-trial evidence of multiple EEG components involved in decision-making..." (line 64). Many readers would disagree with this. For example, Philiastides, Ratcliff, & Sadja (2006) use a single-trial analysis to unravel early and late EEG components relating to decision difficulty and accuracy (across different perceptual decisions), which could be related to the components in the current work. Other, network-based single-trial EEG analyses (e.g., Si et al., 2020, NeuroImage, Sturn et al., 2016 J Neurosci Methods) could also be related to the current component approach. Yet other approaches have used inverse encoding models to examine EEG components related to separable decision processes within trials (e.g., Salvador et al., 2022, Nat Comms). The results of the current work are consistent with this previous work - the two components from Philiastides et al., 2006 can be mapped onto the components in the current work, and Salvador et al., 2022 also uncover stimulus- and decision-deliberation related components.
We completely agree with the reviewer that the link to previous work was insufficient. We now include all references that the reviewer points out both in the introduction (see response R3.2) and in the discussion (see response R3.4). We wish to thank the reviewer for bringing these papers to our attention as they are important for the manuscript.
The authors relate their components to ERPs. This prompts the question of whether we would get the same results with ERP analyses (and, on the whole, the results of the current work are consistent with conclusions based on ERP analyses, with the exception of the missing motor component). It's nice that this analysis is single-trial, but many of the follow-up analyses are based on grouping by condition anyway. Even the single-trial analysis presented in Figure 4 could be obtained by median splits (given the hypotheses propose opposite directions of effects, except for the linear model).
We do not agree with the reviewer in the sense that classical ERP analyses would require much more data-points. The performance of the method is here to use the information shared across all contrast levels to be able to model the processing time of a single contrast level (6 trials per participant). Furthermore, as stated in the response to R1.4 and R1.5, the aim of the paper is to have the time of information processing components which cannot be achieved with classical ERPs without strong, and likely false, assumptions.
Medium Comments:
(1) The presentation of Piéron's law for the behavioural analysis is confusing. First, both laws should be clearly defined for readers who may be unfamiliar with this work. I found the proposal that Piéron's law predicts decreasing RT for increasing pedestal contrast in a contrast discrimination paradigm task surprising, especially given the last author's previous work. For example, Donkin and van Maanen (2014) write "However, the commonality ofPiéron's Law across so many paradigms has lead researchers (e.g., Stafford & Gurney, 2004; Van Maanen et al., 2012) to propose that Piéron's Law is unrelated to stimulus scaling, but is a result of the architecture of the response selection (or decision making) process." The pedestal contrast is unrelated to the difficulty of the contrast discrimination task (except for the consideration of Fechner's law). Instead, Piéron's law would apply to the subjective difference in contrast in this task, as opposed to the pedestal contrast. The EEG results are consistent with these intuitions about Piéron's law (or more generally, that contrast is accumulated over time, so a later EEG component for lower pedestal contrast makes sense): pedestal contrast should lead to faster detection, but not necessarily faster discrimination. Perhaps, given the complexity of the manuscript as a whole, the predictions for the behavioural results could be simplified?
We agree that the initial version was confusing. We now clarified the presentation of Piéron's law at the end of the introduction (see also response to R2).
Once Fechner's law is applied, decision difficulty increases with increasing contrast, so Piéron's law on the decision-relevant intensity (perceived difference in contrast) would also predict increasing RT with increasing pedestal contrast. It is unlikely that the data are of sufficient resolution to distinguish a log function from a power of a log function, but perhaps the claim on line 189 could be weakened (the EEG results demonstrate Piéron's law for detection, but do not provide evidence against Piéron's law in discrimination decisions).
This is an excellent observation, thank you for bringing it to our attention. Indeed, the data support the notion that Pieron’s law is related to detection, but do not rule out that it is also related to decision or discrimination. In earlier work, we (Donkin & Van Maanen, 2014) addressed this question as well, and reached a similar conclusion. After fitting evidence accumulation models to data, we found no linear relationship between drift rates and stimulus difficulty, as would have been the case if Pieron's law could be fully explained by the decision process (as -indirectly- argued by Stafford & Gurney, 2004; Van Maanen et al., 2012). The fact that we observed evidence for a non-linear relationship between drift rates and stimulus difficulty led us to the same conclusion, that Pieron’s law could be reflected in both discrimination and decision processes. We added the following comment to the discussion about the functional locus of Pieron's law to clarify this point:
“Note that this observation is not necessarily incongruent with theoretical work that argued that Piéron’s law could also be a result of a response selection mechanism (Stafford and Gurney, 2004; Van Maanen et al., 2012; Palmer et al., 2005). It could be that differences in stimulus intensity between the two options also contribute to a Piéron like relationship in the later intervals, that is convoluted with Fechner’s law (see Donkin and Van Maanen, 2014, for a similar argument). Unfortunately, our data do not allow us to discriminate between a pure logarithmic growth function and one that is mediated by a decreasing power function”.
(2) Appendix 1 shows that the event detection of the HMP method will also pick up on ramping activity. The description of the problem in the introduction is that event-like activity could look like ramping when averaged across trials. To address this problem, the authors should simulate events (with some reasonable dispersion in timing such that they look like ramping when averaged) and show that the HMP method would not pull out something that looked like ramping. In other words, the evidence for ramping in this work is not affected by the previously identified confounds.
We agree that this demonstration was necessary and thus added the suggested simulation to Appendix 1. As can be seen in the Figure 1 of the appendix, when we simulate a half-sine the average ERP based on the timing of the event looks like a half-sine.
(3) Some readers may be interested in a fuller discussion of the failure of the Fechner diffusion model in the speed condition.
We are unsure which failure the reviewer refers to but assumed it was in relation to the behavioral results and thus added:
It is unlikely that neither Piéron nor Fechner law impact the RT in the speed condition. Instead this result is likely due to the composite nature of the RT where both laws co-exist in the RT but cancel each other out due to their opposite prediction.
Minor Comments:
(1) "By-trial" is used throughout. Normally, it is "trial-by-trial" or "single-trial" or "trial-wise".
We replaced all occurrences of “by-trial” with the three terms suggested were appropriate.
(2) Line 22: "The sum of the times required for the completion of each of these precessing steps is the reaction time (RT)." The total time required. Processing.
Corrected for both.
(3) Line 26/27: "Despite being an almost two century old problem (von Helmholtz, 2021)." Perhaps the citation with the original year would make this point clearer.
We agree and replaced the citation.
(4) Line 73: "accounted by estimating". Accounted for by estimating.
Corrected.
(5) Line 77 "provides an estimation on the." Of the.
Corrected.
(6) Line 86: "The task of the participants was to answer which of two sinusoidal gratings." The picture looks like Gabor's? Is there a 2d Gaussian filter on top of the grating? Clarify in the methods, too.
We incorrectly described the stimuli as those were indeed just Gabor’s. This is now corrected both in the main text and the method section.
(7) Figure 1 legend: "The Fechner diffusion law" Fechner's law or your Fechner diffusion model?
Law was incorrect so we changed to model as suggested.
(8) Line 115: "further allows to connects the..." Allows connecting the.
Corrected.
(9) Line 123: "lower than 100 ms or higher than..." Faster/slower.
Corrected.
(10) Line 131: "To test what law." Which law.?
Corrected to model.
(11) Figure 2 legend: "Left: Mean RT (dot) and average fit (line) over trials and participants for each contrast level used." The fit is over trials and participants? Each dot is? Average trials for each contrast level in each participant?
This sentence was corrected to “Mean RT (dot) for each contrast level and averaged predictions of the individual fits (line) with Accuracy (Top) and Speed (Bottom) instructions.”.
(12) Line 231: "A comprehensive analysis of contrast effect on". The effect of contrast on.
This title was changed to “functional interpretation of the events”.
(13) Line 23: "the three HMP event with". Three HMP events.
The sentence no longer exists in the revised manuscript.
(14) Line 270: "Secondly, we computed the Pearson correlation coefficient between the contrast averaged proportion of correct." Pearson is for continuous variables. Proportion correct is not continuous. Use Spearman, Kendall, or compute d'.
The reviewer rightly pointed out our error, we corrected this by computing Spearman correlation.
(15) Line 377: "trial 𝑛 + 1 was randomly sampled from a uniform distribution between 0.5 and 1.25 seconds." It's just confusing why post-response activity in Figure 5 does look so consistent. Throughout methods: "model was fitted" should be "was fit", and line 448, "were split".
We do not have a specific hypothesis of why the post-response activity in the previous Figure 5 was so consistent. Maybe the Gaussian window (same as in other manuscripts with a similar figure, e.g. O’Connell et al. 2012) generated this consistency. We also corrected the errors mentioned in the methods.
(16) The linear mixed models paragraph is a bit confusing. Can it clearly state which data/ table is being referred to and then explain the model? "The general linear mixed model on proportion of correct responses was performed using a logit link. The linear mixed models were performed on the raw milliseconds scale for the interval durations and on the standardized values for the electrode match." We go directly from proportion correct to raw milliseconds...
The confusion was indeed due to the initial inclusion of a general linear mixed model on proportion correct which was removed as it was not very informative. The new revision should be clearer on the linear mixed models (see first sentence of subsection ‘linear mixed models' in the method section).
(17) A fuller description of the HMP model would be appreciated.
We agree that this was necessary and added the description of the HMP model in the corresponding method section “Hidden multivariate pattern” in addition to a more comprehensive presentation of HMP in the first paragraph of the Result and Discussion sections.
(18) Line 458: "Fechner's law (Fechner, 1860) states that the perceived difference (𝑝) between the two patches follows the logarithm of the difference in physical intensity between..." ratio of physical intensity.
Corrected.
(19) P is defined in equations 2 and 4. I would include the beta in equation 4, like in equation 2, then remove the beta from equations 3 and 5 (makes it more readable). I would also just include the delta in equation 2, state that in this case, c1 = c+delta/2 or whatever.
This indeed makes the equation more readable so we applied the suggestions for equations 2, 3, 4 and 5. The delta was not added in equation 2 but instead in the text that follows:
“Where 𝐶1 = 𝐶0 + 𝛿, again with a modality and individual specific adjustment slope (𝛽).”
(20) The appendix suggests comparing the amplitudes with those in Figure 3, but the colour bar legend is missing, so the reader can only assume the same scale is used?
We added the color bar as it was indeed missing. Note though that the previous version displayed the estimation for the simulated data while this plot in the revised manuscript shows the solution on real data obtained after downsampling the data (and therefore look for a larger pattern as in the main text). We believe that this representation is more useful given that the solution for the downsampled data is no longer the same as the one in the main text (due to the difference in pattern width).
-
-
spitfirenews.com spitfirenews.com
-
“You people would ‘it’s not okay to comment on women’s bodies and she’s always been skinny’
I believe that "Skinny" as a beauty standard is just a leftover effect of Gay Men of Power in the Designer Fashion industry choosing women that look like the men they want to fuck
-
-
www.biorxiv.org www.biorxiv.org
-
Note: This response was posted by the corresponding author to Review Commons. The content has not been altered except for formatting.
Learn more at Review Commons
Reply to the reviewers
We are grateful to the reviewers for their thoughtful and constructive evaluations of our manuscript. Their comments helped us clarify key aspects of the study and strengthen both the presentation and interpretation of our findings. The central goal of this work is to dissect how the opposing activities of GATA4 and CTCF coordinate chromatin topology and transcriptional timing during human cardiomyogenesis. The reviewers’ feedback has allowed us to refine this message and better contextualize our results within the broader framework of chromatin regulation and cardiac development.
In response to the reviews, in our preliminary revision we have already implemented substantial improvements to the manuscript, including additional analyses, clearer data visualization, and revisions to the text to avoid overinterpretation. These refinements enhance the robustness of our conclusions without altering the overall scope of the study. A small number of additional analyses and experiments are ongoing and will be added to the full revision, as detailed below.
We believe that the revised manuscript, together with the planned updates, fully addresses the reviewers’ concerns and substantially strengthens the contribution of this work to the field.
Reviewer 1 – Point 1:
In the datasets you are examining, what are the relative percentages in each of the four groups relating compartmentalization change to expression change (A→B, expression up; A→B, down; B→A, up; B→A, down)?
We quantified compartment–expression relationships using Hi-C and bulk RNA-seq from H9 ESCs and CMs. The percentages for each category are shown below and incorporated into updated Figure S2H.
Group
Downregulated in CM
Upregulated in CM
A-to-A
11.92%
8.44%
A-to-B
18.20%
2.79%
B-to-A
7.96%
18.07%
B-to-B
14.36%
6.44%
A chi-squared test comparing observed vs. expected distributions (based on gene density across bins) confirmed a strong association between compartment dynamics and transcriptional behavior. B-to-A genes are significantly enriched among genes upregulated in CMs, while A-to-B genes are enriched among those downregulated (updated Figure S2H).
We next assessed with GSEA how these gene classes respond to GATA4 and CTCF knockdown. In 2D CMs, GATA4 knockdown reduces expression of CM-upregulated B-to-A genes and increases expression of CM-downregulated A-to-B genes, whereas CTCF knockdown produces the opposite pattern (updated Figure 2F).
Applying the same analysis to cardioid bulk RNA-seq (updated Figure 4E) revealed the strongest effects in SHF-RV organoids, consistent with monolayer data. In SHF-A organoids, only GATA4 knockdown had a measurable impact on CM-upregulated B-to-A and CM-downregulated A-to-B genes. Because the subsets of CM-downregulated B-to-A and CM-upregulated A-to-B genes were very small and showed no consistent trends, Figure 4 focuses on the two informative categories only. The full classification is provided in Reviewer Figure 1 below.
(The figure cannot be rendered in this text-only format)
Reviewer Figure 1. GSEA for CM-upregulated B-to-A and CM-downregulated A-to-B genes. p-values by Adaptive Monte-Carlo Permutation test.
Reviewer 1 – Point 2
This phrase in the abstract is imprecise: ‘whereas premature CTCF depletion accelerates yet confounds cardiomyocyte maturation.’
The abstract has been revised to: “whereas premature CTCF depletion accelerates yet alters cardiomyocyte maturation.” (lines 29-30).
Reviewer 1 – Point 3
Regarding this statement: "Disruption of [3D chromatin architecture] has been linked to genetic dilated cardiomyopathy (DCM) caused by lamin A/C mutations8,9, and mutations in chromatin regulators are strongly enriched in de novo congenital heart defects (CHD)10, underscoring their pathogenic relevance11." The first studies to implicate chromatin structural changes in heart disease, including the role of CTCF in that process, were PMID: 28802249, a model of acquired, rather than genetic, disease.
We added the following sentence to the paragraph introducing CTCF: “Moreover, depletion of CTCF in the adult cardiomyocytes leads to heart failure28,29.” (line 72)
Reviewer 1 – Point 4
Can you quantify this statement: ‘the compartment switch coincided with progressive reduction of promoter–gene body interactions’?
We quantified promoter–gene body contacts by calculating the area under the curve (AUC) of the virtual 4C signal derived from H9 Hi-C data across differentiation. As a result of this analysis we added the following sentence: “Quantitatively, interactions between the TTN promoter and its gene body decreased by ~55% from the pluripotent stage to day 80 cardiomyocytes.” (lines 89-91).
Reviewer 1 – Point 5
Regarding this statement: "six regions became less accessible in CMs, correlating with ChIP-seq signal for the ubiquitous architectural protein CTCF." I don't see 6 ATAC peaks in either TTN trace in Figure 1A.
We corrected the text as it follows: “TTN experienced clear changes in chromatin accessibility during CM differentiation: ATAC-seq identified two CM-specific peaks that correlated with ChIP-seq signal for the cardiac pioneer TF GATA4 at the two promoters, one driving full length titin and the other the shorter cronos isoform. In contrast, two regions became less accessible in CMs, correlating with two of the six ChIP-seq peaks for the ubiquitous architectural protein CTCF” (lines 93-97). We attribute the differences between ChIP-seq and ATAC-seq profiles to methodological sensitivity and/or biological variability between datasets generated in different laboratories and cell batches.
Reviewer 1 – Point 6
Western blots need molecular weight markers.
We edited the relevant panels accordingly (updated Figures 1E and 2B).
Reviewer 1 – Point 7
Regarding this statement: "The decrease in CTCF protein levels may explain its selective detachment from TTN during cardiomyogenesis." At face value, these findings suggest the opposite: i.e. that a massive downregulation of CTCF at protein level should affect its binding across the genome, which is not tested and is hard to evaluate between ChIP-seq studies from different groups and from different developmental timeframes.
We revised the text to avoid implying selective detachment and performed a genome-wide analysis of CTCF occupancy using ENCODE ChIP-seq datasets generated by the same laboratory with matched protocols in hESCs and hESC-derived CMs. This analysis shows that 43.2% of CTCF sites present in ESCs are lost in CMs, whereas only 5.7% are gained, confirming a broad reduction in CTCF binding during differentiation. These results are now included in__ updated Figure 1B__.
Reviewer 1 – Point 8a
A couple thoughts on the FISH experiments in Figure 2. A claim of 'impaired B-A transition' would be more convincing if you show, by FISH, that the relative distance of TTN from lamin B increases with differentiation.
Although prior work from us and others has established that TTN transitions from the nuclear periphery in hESCs to a more internal position during cardiomyogenesis (Poleshko et al. 2017; Bertero et al. 2019a), we are reproducing this trajectory in WTC11 hiPSCs as part of the FISH experiments for the full revision.
__Reviewer 1 – Point 8b __
In the [FISH] images: are you showing a total projection of all z planes? One assumes the quantitation is relative to a 3D reconstruction in which the lamin B signal is restricted to the periphery. Have you shown this? __
Quantification was performed on full 3D reconstructions from Z-stacks, as detailed in the Methods (lines 721-727). While the original submission displayed maximum-intensity projections, updated Figure 2D and Figure S2E now show representative single optical sections, which more clearly highlight the spatial relationship between the TTN locus and the nuclear lamina.
Reviewer 1 – Point 8c
Lastly, these data are very interesting and important, provoking reexamination of your interpretation of the results in Figure 1. Figure 1 was interpreted to show that less CTCF binding led to decreased lamina (and thus B compartment) association during development. Figure 2 shows that depleting CTCF does not change association of TTN with lamina.
Our interpretation is that by day 25 of hiPSC-CM differentiation the TTN locus may have reached its maximal radial repositioning even in control cells, limiting the ability to detect earlier effects of CTCF depletion. To test whether CTCF knockdown accelerates lamina detachment at earlier stages, we are repeating the FISH analysis for the inducible CTCF knockdown line at multiple time points during differentiation.
Reviewer 1 – Point 9
A thought about this statement: "Altogether, these results suggest that GATA4 and CTCF function as positive and negative regulators of B-to-A compartment switching, likely acting through global and local chromatin remodeling, respectively." GATA4 induces TTN expression and its knockdown prevents TTN expression-the evidence that GATA4 affects compartmentalization is unclear. By activating the gene, GATA4 may shift TTN to B classification.
Our current data do not allow us to disentangle whether GATA4-driven transcriptional activation precedes or follows the B-to-A compartment shift. We have therefore removed the mechanistic speculation from this sentence to avoid overinterpretation. Nevertheless, the analyses in updated Figure 2F, discussed in the response to Reviewer 1 - Point 1, show that GATA4 knockdown preferentially reduces expression of CM-upregulated B-to-A genes, while CTCF knockdown has the opposite effect, supporting the conclusion that both factors influence the transcriptional programs associated with B-to-A transitions.
Reviewer 1 – Point 10
__I'm not sure what I am looking at in Figure 3C. Are those traces integration of interactions over a defined window? "Each [mutant is] clearly different from WT" is not obvious from the presentation. The histograms are plotting AUC of what? Interactions of those peaks with the mutated region? I genuinely appreciate how laborious this experiment must have been and encourage you to explain better what you are showing. __
We revised the main text to avoid overstating the differences (“clearly” “in a similar manner”, line 192) and expanded the l__egends of updated Figures 3C–D__ to clarify what is being shown: “(C) 4C-seq in hiPSCs using the promoter-proximal region of TTN as viewpoint. The top panel shows raw interaction profiles. The lower panels plot pairwise differences between conditions to reveal subtle changes. A schematic indicating the 4C viewpoint is included for clarity. Right inset: zoom of the CBS4–5 region. Mean of n = 3 cultures. (D) AUC of the differential 4C-seq signal for defined intervals (panel C). p-values by one-sample t-test against μ = 0.”. We also added a visual cue in updated Figure 3C indicating the 4C viewpoint to facilitate interpretation.
Reviewer 1 – Point 11
Again acknowledging how challenging these experiments are: when you mutant a locus, you change CTCF binding but you also change the DNA. Thus, attributing the changes in interactions to presence/absence of CTCF binding is difficult, because the DNA substrate itself has changed. Perhaps you are presenting all of this as a negative result, given the modest effect on transcription, which is as important as a positive result, given the assumptions usually made about such things. But the results are not clearly described and your interpretation seems to go between implying the structural change causative and being agnostic.
We recognize that deleting a genomic region can affect both CTCF binding and the DNA substrate itself. For this reason, we implemented two parallel genome-editing strategies:
(1) a straightforward Cas9-mediated deletion of ~100 bp centered on each CBS, and
(2) a more precise HDR approach replacing only the 20 bp core CTCF motif.
Because the HDR strategy succeeded, all downstream analyses were carried out on these minimal edits, which substantially limit disruption of other transcription factor motifs and reduce the likelihood of sequence-dependent polymer effects unrelated to CTCF.
Nevertheless, to avoid implying unwarranted causality in the absence of more conclusive evidence, we added a paragraph to the Discussion outlining these limitations, including the sentence: “Our study also reflects general challenges in separating chromatin-architectural and transcriptional mechanisms. Although the CBS edits were restricted to the core CTCF motifs, additional sequence-dependent effects cannot be fully excluded, and we therefore interpret the resulting changes as consistent with—but not exclusively due to—loss of CTCF binding.” (lines 365-368)
Reviewer 1 - Point 12.
Figure 4C: since you have RNA-seq data, a much more objective way to present these data would be to show all data (again, A-B, up; A-B, down; B-A, up; B-A, down) and the effects of CTCF or GATA4. Regardless, you can still focus on the cardiac specific genes. But my guess is if you examine all genes, the pattern you show in panel C will not be present in the majority of cases. Furthermore, if this hypothesis is wrong, such an analysis will allow you to identify other genes affected by the mechanisms you describe and your analysis will test whether these mechanisms are in fact conserved at different loci.
As outlined in our response to Point 1, we extended the analysis to all genes undergoing compartment changes and incorporated this into the cardioid RNA-seq dataset. This revealed a clear and consistent relationship between GATA4 or CTCF knockdown and the expression of B-to-A and A-to-B gene classes (updated Figure 4E).
Reviewer 2 - Point 1.1
1. CTCF regulation at TTN locus:
(1) Figure 1A: The claim of the authors about convergent CTCF sites and transcriptional activation of TTN is quite simplistic. This claim is only valid when we know where cohesin is loaded. If cohesin is loaded at then intragenic GATA4 binding site, then the only important CTCF sites is at the promoter of TTN. I suggest that the authors read few more publications which may help the authors to better understand how cohesin and CTCF team up to regulate transcription, such as Hsieh et al., Nature Genetics, 2022; Liu et al., Nature Genetics, 2021; Rinzema et al., Nature Structural and Molecular Biology, 2022.
__Suggestion: The authors should add cohesin (RAD21/SMC1A) and NIPBL ChIP-seq for better interpretation. __
In line with the reviewer’s insightful suggestion, we integrated cohesin ChIP-seq data into updated Figure 1A. Specifically, we added a RAD21 ChIP-seq track from hESCs, which provides direct evidence of cohesin occupancy across the TTN locus. RAD21 binding closely parallels CTCF binding at five sites within the gene body, supporting a model in which promoter-proximal CTCF anchors cohesin to stabilize repressive loops at this locus. This analysis substantially strengthens the mechanistic framework and is consistent with the studies recommended by the reviewer, which we have now cited (lines 68 and 104).
Reviewer 2 - Point 1.2. (2) Figure 3B: If delta2CBS only has heterozygenous deletion of CBS6, why we would expect the binding will be weaken to 50%. However, the CTCF binding is reduced to around 1/10 in the ChIP-qPCR. How do the authors explain this?
Sequencing of the Δ2CBS line shows that one CBS6 allele carries the intended EcoRI replacement, while the second allele contains a 2-bp deletion within the core CTCF motif (Figure S3C). Remarkably, this small deletion is sufficient to abolish CTCF binding, resulting in complete loss of occupancy at CBS6 despite heterozygosity. We clarified this in the text as follows: “CTCF ChIP-qPCR in hiPSCs confirmed complete loss of CTCF binding at the targeted sites, including CBS6 in the Δ2CBS line, indicating that the 2-bp deletion sufficed to disrupt CTCF binding while occupancy at other CBSs remained unaffected.” (lines 187–189).
Reviewer 2 - Point 1.3a (3) Figure 3C: There are two problems with the 4C experiments: (a) The changes are really mild. In fact, none of the p-values in Figure 3D are significant.
The effect of deleting CBS1 is indeed modest, consistent with reports that individual CTCF binding sites often show functional redundancy (i.e., Rodríguez-Carballo et al. 2017; Barutcu et al. 2018; Kang et al. 2021). Nevertheless, our 4C-seq experiments have reproducibly shown the same directional trend across biological replicates. To increase statistical power and more rigorously assess the robustness of this effect, we are generating additional 4C replicates as part of the full revision.
Reviewer 2 - Point 1.3b [In the 4C experiments] (b) The authors should also consider a model that CTCF directly serves as a repressor. In this way, 3D genome may not be involved. B-A switch is simply caused by the activation of the locus.
We now explicitly acknowledge this possibility in the Discussion. The revised text states: “Moreover, our data cannot unambiguously separate CTCF’s architectural role from potential direct repressive activity. Both mechanisms could contribute to the observed effects, and our findings likely reflect the combined influence of CTCF on chromatin topology and gene regulation.” (lines 368–371).
Reviewer 2 - Point 2.1a 2. __(CTCF) detachment: The authors mentioned few times "detachment". In the context of this manuscript, the authors indicate detachment from nuclear lamina. However, the authors haven't provide convincing evidence about this. __
In the two instances where we used the term “detachment,” we intended it to refer exclusively to reduced CTCF binding to DNA, not to lamina repositioning. To avoid ambiguity, we have replaced “detachment” with “reduced binding” in both locations (lines 123 and 329). We do not use this term to describe TTN–lamina positioning.
Reviewer 2 - Point 2.1b (1) Figure 1D: I doubt whether such changes of CTCF protein abundance will lead to LAD detachment. Suggest the authors read van Schaik et al., Genome Biology, 2022. With the full depletion of CTCF, the effects on LADs are still very restricted.
We agree that the observed correlation between reduced CTCF levels and the relocation of TTN away from a LAD does not establish causality. As outlined in our response to Reviewer 1 – Point 8c, we are performing additional FISH experiments at earlier differentiation stages in the CTCF inducible knockdown line to directly assess whether partial CTCF depletion is sufficient to alter the timing of TTN–lamina separation.
Reviewer 2 - Point 2.2 (2) Figure 2D: Lamin B1 should be mostly at nuclear periphery. I have few questions: (1) is the antibody specific? (2) do these cells carry mutation in LMNB1 gene? (3) is the staining actually LMNA?
As also clarified in response to Reviewer 1 – Point 8b, the original images displayed maximum-intensity projections of Z-stacks, which obscured the peripheral distribution of LMNB1. We have updated Figure 2D and Figure S2E to show representative individual optical sections, which more clearly display the expected peripheral LMNB1 signal. We also confirm that the antibody used is specific for LMNB1 and previously validated (Bertero et al. 2019b), and that the WTC11-derived lines used in this study carry no mutation in LMNB1.
Reviewer 2 - Point 3
3. Opposite functions of GATA4 and CTCF: These data in Figure 5E-H argues the opposite role of GATA4 and CTCF in transcriptional regulation. Would it be that CTCF KD just affected cell proliferation, which is actually known for many cell types, rather than affect CM differentiation process? If this is the reason, inversed correlation between CTCF KD and GATA4 KD in Figure 4D could also be explained by opposite effects on cell cycle.
We directly evaluated this possibility. In FHF–LV cardioids, cell cycle profiling in Figure 6C and Figure S6C (now S7C) showed that CTCF knockdown does not alter the distribution of CMs across G1/S/G2–M phases, in contrast to the marked increase in proliferation observed with GATA4 knockdown.
Because this comment referred specifically to the SHF data, we also analyzed mitotic gene expression in the SHF–RV bulk RNA-seq dataset using GSEA. CTCF knockdown did not significantly enrich any cell cycle–related gene sets, whereas GATA4 knockdown produced a strong enrichment for mitotic cell cycle terms, in line with FHF-LV data (Reviewer Figure 2).
These results are summarized in updated Figure S5C, reporting also the results of the broader GSEA analysis, and together indicate that the transcriptional divergence between CTCF and GATA4 knockdown is not simply explained by opposing effects on proliferation.
(The figure cannot be rendered in this text-only format)
Reviewer Figure 2. GSEA for mitotic cell cycle in SHF-RV after inducible knockdown of CTCF (left) or GATA4 (right). p-values by Adaptive Monte-Carlo Permutation test.
Reviewer 2 - Point 4 4. In discussion, the authors suggested that CTCF is a local chromatin remodeller. In my view, association with local chromatin compaction doesn't qualify CTCF as a chromatin remodeler. To my knowledge, CTCF does not have an enzymatic domain, then how does it remodel chromatin?
Our intended meaning was that CTCF shapes 3D chromatin architecture through its role in organizing intergenic looping, not that it remodels chromatin enzymatically. To avoid confusion, we have removed the original sentence from the Discussion.
Reviewer 2 - Point 5. 5. Some conclusions are drawn based on insignificant p-values, e.g. Figure 2F, Figure 3D, etc. The authors should be careful about their conclusion, and tone down their statement for the observations have borderline significance.
The conclusions based on bulk RNA-seq have been revised in response to Reviewer 1 – Point 1 (updated Figure 2F). By subsetting B-to-A and A-to-B genes according to their expression dynamics, this analysis now yields clearer and statistically significant differences between conditions.
Regarding the 4C-seq data, as acknowledged in Reviewer 2 – Point 3a, the observed effects are modest. We are generating additional biological replicates to increase statistical power. In the meantime, we have adjusted the text to avoid overstating these findings. The revised manuscript now states: “While the difference did not reach significance, these trends suggest …” (lines 199–200).
Reviewer 2 - Minor comment 1. Minor comments: 1. Figure 1A: (1) I suggest to label two promoters in the gene model. It's unclear in the figure in the current version; (2) I was a bit confused with the way how the authors labeled CTCF directionality. I thought there are a lot of promoters. Why didn't they use triangles?
We updated Figure 1A to label both TTN promoters and indicate their orientation. For CTCF sites, we now clearly display the motif direction and core binding region as determined by FIMO analysis of the CTCF ChIP-seq peaks, improving consistency and interpretability.
Reviewer 2 - Minor comment 2. 2. Figure 2C: I think the drastical reduction of titin-mEGFP levels is only due to the way how the authors analyze their FACS data. Can the author quantify on median fluorescence intensity?
The gating strategy for titin-mEGFP⁺ cells was defined using a reporter-negative control, and cells lacking TNNT2 expression showed no detectable titin-mEGFP signal, confirming the specificity of the gate. To complement this analysis, we also quantified the median fluorescence intensity (MFI) of titin-mEGFP⁺ cells. The MFI analysis corroborates the original findings, showing a significant decrease in GATA4 knockdown and an increase in CTCF knockdown (updated Figure S2D).
__Reviewer 2 - Minor comment 3. 3. Figure S2G: P value should be -log10, I assume. Please label it accurately. __
We appreciate the reviewer pointing out this labeling error. In the revised manuscript, this panel has been removed to accommodate the updated compartment–expression analysis now presented in updated Figure 2H (see response to Reviewer 1 – Point 1), and the issue is no longer applicable.
References
Barutcu AR, Maass PG, Lewandowski JP, Weiner CL, Rinn JL. 2018. A TAD boundary is preserved upon deletion of the CTCF-rich Firre locus. Nat Commun 9: 1444.
Bertero A, Fields PA, Ramani V, Bonora G, Yardımcı GG, Reinecke H, Pabon L, Noble WS, Shendure J, Murry CE. 2019a. Dynamics of genome reorganization during human cardiogenesis reveal an RBM20-dependent splicing factory. Nature communications 10: 1538.
Bertero A, Fields PA, Smith AS, Leonard A, Beussman K, Sniadecki NJ, Kim D-H, Tse H-F, Pabon L, Shendure J, et al. 2019b. Chromatin compartment dynamics in a haploinsufficient model of cardiac laminopathy. Journal of Cell Biology 218: 2919–44.
Kang J, Kim YW, Park S, Kang Y, Kim A. 2021. Multiple CTCF sites cooperate with each other to maintain a TAD for enhancer–promoter interaction in the β-globin locus. The FASEB Journal 35: e21768.
Poleshko A, Shah PP, Gupta M, Babu A, Morley MP, Manderfield LJ, Ifkovits JL, Calderon D, Aghajanian H, Sierra-Pagán JE, et al. 2017. Genome-Nuclear Lamina Interactions Regulate Cardiac Stem Cell Lineage Restriction. Cell 171: 573–587.
Rodríguez-Carballo E, Lopez-Delisle L, Zhan Y, Fabre PJ, Beccari L, El-Idrissi I, Huynh THN, Ozadam H, Dekker J, Duboule D. 2017. The HoxD cluster is a dynamic and resilient TAD boundary controlling the segregation of antagonistic regulatory landscapes. Genes Dev 31: 2264–2281.
-
Note: This preprint has been reviewed by subject experts for Review Commons. Content has not been altered except for formatting.
Learn more at Review Commons
Referee #2
Evidence, reproducibility and clarity
Becca et al. characterized the functions of GATA4 and CTCF in the context of cardiomyogenesis. The authors aim to establish a link between 3D genome changes (A/B compartment and long-range chromatin interactions) and activation of cardiac specific genes such as TTN. They showed opposite effects of GATA4 and CTCF in regulating these genes as well as phenotypical traits. I have the following suggestions and questions:
Major comments:
- CTCF regulation at TTN locus:
(1) Figure 1A: The claim of the authors about convergent CTCF sites and transcriptional activation of TTN is quite simplistic. This claim is only valid when we know where cohesin is loaded. If cohesin is loaded at then intragenic GATA4 binding site, then the only important CTCF sites is at the promoter of TTN. I suggest that the authors read few more publications which may help the authors to better understand how cohesin and CTCF team up to regulate transcription, such as Hsieh et al., Nature Genetics, 2022; Liu et al., Nature Genetics, 2021; Rinzema et al., Nature Structural and Molecular Biology, 2022.
Suggestion: The authors should add cohesin (RAD21/SMC1A) and NIPBL ChIP-seq for better interpretation. (2) Figure 3B: If delta2CBS only has heterozygenous deletion of CBS6, why we would expect the binding will be weaken to 50%. However, the CTCF binding is reduced to around 1/10 in the ChIP-qPCR. How do the authors explain this?
(3) Figure 3C: There are two problems with the 4C experiments: (a) The changes are really mild. In fact, none of the p-values in Figure 3D are significant; (b) The authors should also consider a model that CTCF directly serves as a repressor. In this way, 3D genome may not be involved. B-A switch is simply caused by the activation of the locus. 2. (CTCF) detachment: The authors mentioned few times "detachment". In the context of this manuscript, the authors indicate detachment from nuclear lamina. However, the authors haven't provide convincing evidence about this.
(1) Figure 1D: I doubt whether such changes of CTCF protein abundance will lead to LAD detachment. Suggest the authors read van Schaik et al., Genome Biology, 2022. With the full depletion of CTCF, the effects on LADs are still very restricted.
(2) Figure 2D: Lamin B1 should be mostly at nuclear periphery. I have few questions: (1) is the antibody specific? (2) do these cells carry mutation in LMNB1 gene? (3) is the staining actually LMNA? 3. Opposite functions of GATA4 and CTCF: These data in Figure 5E-H argues the opposite role of GATA4 and CTCF in transcriptional regulation. Would it be that CTCF KD just affected cell proliferation, which is actually known for many cell types, rather than affect CM differentiation process? If this is the reason, inversed correlation between CTCF KD and GATA4 KD in Figure 4D could also be explained by opposite effects on cell cycle. 4. In discussion, the authors suggested that CTCF is a local chromatin remodeller. In my view, association with local chromatin compaction doesn't qualify CTCF as a chromatin remodeler. To my knowledge, CTCF does not have an enzymatic domain, then how does it remodel chromatin? 5. Some conclusions are drawn based on insignificant p-values, e.g. Figure 2F, Figure 3D, etc. The authors should be careful about their conclusion, and tone down their statement for the observations have borderline significance.
Minor comments:
- Figure 1A: (1) I suggest to label two promoters in the gene model. It's unclear in the figure in the current version; (2) I was a bit confused with the way how the authors labeled CTCF directionality. I thought there are a lot of promoters. Why didn't they use triangles?
- Figure 2C: I think the drastical reduction of titin-mEGFP levels is only due to the way how the authors analyze their FACS data. Can the author quantify on median fluorescence intensity?
- Figure S2G: P value should be -log10, I assume. Please label it accurately.
Significance
Strengths and limitations:
I feel that single-cell analysis and functional analysis of GATA4 and CTCF using cardiac organoid model are elegant. However, the weak part of the manuscript is the link between 3D genome and activation of TTN. I also think the authors should include more possible explanations for the interpretation of some genome organization data (CTCF site deletion, 4C, etc).
Advance: The study does provide useful information to understand transcriptional regulation during cardiac lineage specification. The link between 3D genome and cardiac lineage specification is conceptually nice but needs more data to support.
Audience: developmental biologists who is interested in heart development and molecular biologists with specific interests in gene regulation.
-
-
arxiv.org arxiv.org
-
Reviewer #1 (Public review):
Summary:
The authors report the results of a tDCS brain stimulation study (verum vs sham stimulation of left DLPFC; between-subjects) in 46 participants, using an intense stimulation protocol over 2 weeks, combined with an experience-sampling approach, plus follow-up measures after 6 months.
Strengths:
The authors are studying a relevant and interesting research question using an intriguing design, following participants quite intensely over time and even at a follow-up time point. The use of an experience-sampling approach is another strength of the work.
Weaknesses:
There are quite a few weaknesses, some related to the actual study and some more strongly related to the reporting about the study in the manuscript. The concerns are listed roughly in the order in which they appear in the manuscript.
(1) In the introduction, the authors present procrastination nearly as if it were the most relevant and problematic issue there is in psychology. Surely, procrastination is a relevant and study-worthy topic, but that is also true if it is presented in more modest (and appropriate) terms. The manuscript mentions that procrastination is a main cause of psychopathology and bodily disease. These claims could possibly be described as 'sensationalized'. Also, the studies to support these claims seem to report associations, not causal mechanisms, as is implied in the manuscript.
(2) It is laudable that the study was pre-registered; however, the cited OSF repository cannot be accessed and therefore, the OSF materials cannot be used to (a) check the preregistration or to (b) fill in the gaps and uncertainties about the exact analyses the authors conducted (this is important because the description of the analyses is insufficiently detailed and it is often unclear how they analyzed the data).
(3) Related to the previous point: I find it impossible to check the analyses with respect to their appropriateness because too little detail and/or explanation is given. Therefore, I find it impossible to evaluate whether the conclusions are valid and warranted.
(4) Why is a medium effect size chosen for the a priori power analysis? Is it reasonable to assume a medium effect size? This should be discussed/motivated. Related: 18 participants for a medium effect size in a between-subjects design strikes me as implausibly low; even for a within-subjects design, it would appear low (but perhaps I am just not fully understanding the details of the power analysis).
(5) It remains somewhat ambiguous whether the sham group had the same number of stimulation sessions as the verum stimulation group; please clarify: Did both groups come in the same number of times into the lab? I.e., were all procedures identical except whether the stimulation was verum or sham?
(6) The TDM analysis and hyperbolic discounting approach were unclear to me; this needs to be described in more detail, otherwise it cannot be evaluated.
(7) Coming back to the point about the statistical analyses not being described in enough detail: One important example of this is the inclusion of random slopes in their mixed-effects model which is unclear. This is highly relevant as omission of random slopes has been repeatedly shown that it can lead to extremely inflated Type 1 errors (e.g., inflating Type 1 errors by a factor of then, e.g., a significant p value of .05 might be obtained when the true p value is .5). Thus, if indeed random slopes have been omitted, then it is possible that significant effects are significant only due to inflated Type 1 error. Without more information about the models, this cannot be ruled out.
(8) Related to the previous point: The authors report, for example, on the first results page, line 420, an F-test as F(1, 269). This means the test has 269 residual degrees of freedom despite a sample size of about 50 participants. This likely suggests that relevant random slopes for this test were omitted, meaning that this statistical test likely suffers from inflated Type 1 error, and the reported p-value < .001 might be severely inflated. If that is the case, each observation was treated as independent instead of accounting for the nestedness of data within participants. The authors should check this carefully for this and all other statistical tests using mixed-effects models.
(9) Many of the statistical procedures seem quite complex and hard to follow. If the results are indeed so robust as they are presented to be, would it make sense to use simpler analysis approaches (perhaps in addition to the complex ones) that are easier for the average reader to understand and comprehend?
(10) As was noted by an earlier reviewer, the paper reports nearly exclusively about the role of the left DLPFC, while there is also work that demonstrates the role of the right DLPFC in self-control. A more balanced presentation of the relevant scientific literature would be desirable.
(11) Active stimulation reduced procrastination, reduced task aversiveness, and increased the outcome value. If I am not mistaken, the authors claim based on these results that the brain stimulation effect operates via self-control, but - unless I missed it - the authors do not have any direct evidence (such as measures or specific task measures) that actually capture self-control. Thus, that self-control is involved seems speculation, but there is no empirical evidence for this; or am I mistaken about this? If that is indeed correct, I think it needs to be made explicit that it is an untested assumption (which might be very plausible, but it is still in the current study not empirically tested) that self-control plays any role in the reported results.
(12) Figures 3F and 3H show that procrastination rates in the active modulation group go to 0 in all participants by sessions 6 and 7. This seems surprising and, to be honest, rather unlikely that there is absolutely no individual variation in this group anymore. In any case, this is quite extraordinary and should be explicitly discussed, if this is indeed correct: What might be the reasons that this is such an extreme pattern? Just a random fluctuation? Are the results robust if these extreme cells are ignored? The authors remove other cells in their design due to unusual patterns, so perhaps the same should be done here, at least as a robustness check.
(13) The supplemental materials, unfortunately, do not give more information, which would be needed to understand the analyses the authors actually conducted. I had hoped I would find the missing information there, but it's not there.
In sum, the reported/cited/discussed literature gives the impression of being incomplete/selectively reported; the analyses are not reported sufficiently transparently/fully to evaluate whether they are appropriate and thus whether the results are trustworthy or not. At least some of the patterns in the results seem highly unlikely (0 procrastination in the verum group in the last 2 observation periods), and the sample size seems very small for a between-subjects design.
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public review):
Summary:
Biomolecular condensates are an essential part of cellular homeostatic regulation. In this manuscript, the authors develop a theoretical framework for the phase separation of membrane-bound proteins. They show the effect of non-dilute surface binding and phase separation on tight junction protein organization.
Strengths:
It is an important study, considering that the phase separation of membrane-bound molecules is taking the center stage of signaling, spanning from immune signaling to cell-cell adhesion. A theoretical framework will help biologists to quantitatively interpret their findings.
Weaknesses:
Understandably, the authors used one system to test their theory (ZO-1). However, to establish a theoretical framework, this is sufficient.
We acknowledge this limitation. While we agree that additional systems would strengthen the generality of our theory, we note that the focus of this work is to introduce and validate a theoretical framework. As the reviewer notes, this is sufficient for establishing the framework. Nonetheless, we are open to further collaborations or future studies to test the model with other systems.
Reviewer #2 (Public review):
Summary:
The authors present a clear expansion of biophysical (thermodynamic) theory regarding the binding of proteins to membrane-bound receptors, accounting for higher local concentration effects of the protein. To partially test the expanded theory, the authors perform in vitro experiments on the binding of ZO1 proteins to Claudin2 C-terminal receptors anchored to a supported lipid bilayer, and capture the effects that surface phase separation of ZO1 has on its adsorption to the membrane.
Strengths:
(1) The derived theoretical framework is consistent and largely well-explained.
(2) The experimental and numerical methodologies are transparent.
(3) The comparison between the best parameterized non-dilute theory is in reasonable agreement with experiments.
Weaknesses:
(1) In the theoretical section, what has previously been known, compared to which equations are new, should be made more clear.
We have revised the theory section to clearly distinguish previously established formulations from novel contributions following equation (4), which is .
(2) Some assumptions in the model are made purely for convenience and without sufficient accompanying physical justification. E.g., the authors should justify, on physical grounds, why binding rate effects are/could be larger than the other fluxes.
For our problem, binding is relevant together with diffusive transport in each phase. Each process is accompanied by kinetic coefficients that we estimate for the experimental system. For the considered biological systems (and related ones), it is difficult to determine whether other fluxes (see, e.g., Eq. 8(e)) have relaxed or not. We note that their effects are, of course, included in the kinetic model applied to the coarsening of ZO1 surface condensates as boundary conditions. But we cannot exclude that the corresponding kinetic coefficient in the actual biological system is large enough such that, e.g., Eq. (9e) does not vanish to zero “quasi-statically”. We have now added a sentence to the outlook highlighting the relevance of testing those flux-force relationships in biological systems.
(3) I feel that further mechanistic explanation as to why bulk phase separation widens the regime of surface phase separation is warranted.
We have discussed the mechanistic explanation related to bulk protein interaction strength in the manuscript in the section: “Effects of binding affinity and interactions on surface phase separation”. We explained how the bulk interaction parameter affects the binding equilibrium.
(4) The major advantage of the non-dilute theory as compared with a best parameterized dilute (or homogenous) theory requires further clarification/evidence with respect to capturing the experimental data.
We thank reviewer for this helpful question. To address this point, we have added new paragraphs in the conclusion section, which explicitly discuss the necessity of employing the non-dilute theory for interpreting the experimental data.
(5) Discrete (particle-based) molecular modelling could help to delineate the quantitative improvements that the non-dilute theory has over the previous state-of-the-art. Also, this could help test theoretical statements regarding the roles of bulk-phase separation, which were not explored experimentally.
We appreciate the suggestion and agree that such modeling would be valuable. However, this is beyond the scope of the current study.
(6) Discussion of the caveats and limitations of the theory and modelling is missing from the text.
We sincerely appreciate the reviewer’s helpful comment. We have added a discussion in the conclusion section outlining the caveats and limitations of our modeling approach.
Reviewing Editor Comments:
Upon discussing with the reviewers, we feel that this manuscript could significantly be improved if testing the model with a different model system (beyond ZO1/tight junctions), in which case we foresee that we could enhance the strength of evidence from "compelling" to "exceptional". But of course, this is up to the authors to go for it or not, the paper is already very good.
Reviewer #2 (Recommendations for the authors):
(1) Lines 132-134: Re-word, the use of "complex" is confusing.
We have rephrased the sentence for clarity. The revised version reads: ṽ<sub>_𝑃𝑅</sub>_ are the molecular volume and area of the protein-receptor complex ѵ<sub>𝑃𝑅</sub>, respectively”, and the changes have been in the revised manuscript.
(2) Line 154 use of ""\nu"" for volume and area could be avoided for better clarity.
We thank the reviewer for this helpful suggestion. We have removed the statement involving ""\nu"" as these quantities have already been defined in the preceding context.
(3) Line 158 the total "Helmholtz" free energy F...
We have added the word "Helmholtz" to the sentence.
(4) Line 160 typo "In specific,..."
We carefully checked this sentence but could not identify a typo.
(5) For equation 5 explain the physical origins of each term, or provide a reference if this equation is explained elsewhere.
Thank you very much for your valuable suggestions. We have carefully rephrased Equation (5) and added a paragraph immediately afterward to provide a detailed explanation of its physical meaning.
(6) Derivation on lines 163-174 is poorly written. Make the logical flow between the equations clearer.
We greatly appreciate your insightful suggestions. Equation (6) has been carefully revised for clarity, and the explanation has been rewritten to ensure better readability. All modifications are Done.
(7) Define bold "t" in Equation 6.
The variable “t” has been explicitly defined in the context for clarity.
(8) In equations. 7b-7c the nablas (gradients) should be the 2D versions.
We have updated the gradient operators in Equations (7b) and (7c) [Eq. (9) in revised manuscript] to their 2D forms for consistency.
(9) Line 190, avoid referring to the future Equation 14, and state in words what is meant by "thermodynamic equilibrium".
We have added the explanation of “thermodynamic equilibrium” and remove the reference to equation accordingly.
(10) In Equation 11 you don't explain what you are doing ( which is a perturbation around the minimum of the free energy).
We have revised the paragraph before equation (11) [Eq. (13) in revised manuscript] to clarify that the expression represents a perturbation around the minimum of the free energy.
(11) In Equation 12, doesn't this also depend on how you have written equation 6 (not just equation 5).
Eq. (12) [Eq. (14) in revised manuscript] is derived directly from the variation of the total free energy F. In contrast, Eq. (6) contains the time derivative of free energies that were not written in their final form. In the revised version, we have now given the conjugate forces and fluxes in Eqs. (7) and (8) for clarity.
(12) Line 206 specify the threshold of local concentration (or provide a reference).
We have specified the threshold of local concentration in the revised text, and the corresponding statement has been highlighted.
(13) Line 223 is the deviation from ideality captured in a pair-wise fashion? I presume it does not account for N many-body interactions?
Yes, our model is formulated within a mean-field framework that incorporates pairwise (second order) interaction coefficients. For example, 𝜒<sub>𝑃𝑅 -𝑅</sub> characterizes the interaction between the complex 𝑃𝑅 and the free receptor 𝑅, 𝜒<sub>𝑅 -L</sub> the interaction between free receptor 𝑅 and free lipid 𝐿, 𝜒<sub>𝑃𝑅-𝐿</sub> the interaction between complex 𝑃𝑅and free lipid 𝐿. We have stressed this choice of free energy in the revised manuscript.
(14) Line 274, how do the authors know the secondary effects (of which they should mention a few) do not significantly impact the observed behaviour?
We sincerely thank the reviewer for the helpful comment. First, the parameters 𝜒<sub>𝑅 -L</sub> and 𝜒<sub>𝑃𝑅 -𝑅</sub> are not essential based on the experimental observations. For more information, please see our revised paragraph on the choice of the specific parameter values, which has been in the following Eq. (21).
(15) It's not clear how Figures 3 b and c are generated with reference to which parameters are changed to investigate with/without bulk phase separation.
To improve clarity, we have revised Figure 3 to display the corresponding parameter values directly in each panel. Figures 3b and 3c were generated by computing the surface binding curves (as shown in Fig. 2) for each binding affinity 𝜔<sub>𝑃𝑅</sub> and membrane-complex interaction strength 𝜒<sub>𝑃𝑅-𝐿</sub>, under different bulk interaction strengths chi, to compare the cases with and without bulk phase separation.
(16) The jump between theory and the "Mechanism in ..." section is too much. The authors should include the biological context of tight junctions and ZO1 in the main introduction.
We appreciate the reviewer’s suggestion. Following this comment, we have added an extended discussion in the main introduction to provide the necessary biological context of tight junctions and ZO1. In addition, we inserted new bridging paragraphs between the theoretical section and the section “Mechanism in tight junction formation” to create a smoother transition from theory to experiments. These revisions help to better connect the theoretical framework with the biological phenomena discussed in the later section.
-
-
q.utoronto.ca q.utoronto.ca
-
Cindy, on the other hand, is challenged to unmaskand unlearn White settler colonial ways of thinking and being in the world,and to listen and learn from Indigenous people.
I resonate a lot with Cindy. I think trying to unlearn colonial ways can be difficult, but once you're aware of what they look like, it makes it a bit easier. This reminds me of when bell hooks stated, "The enemy within must be transformed before we can confront the enemy out." when referring to internalized sexism (hooks 2014, 12). Broadly speaking, when we talk about internalized sexism, it is all these internalized thoughts that we carry with us about gender norms that may be harmful to us. I think this is quite similar to colonial ways of thinking because, just like internalized sexism, colonial thinking is built into us because it played a role in every step of our lives when growing up in a Western society.
Often, we have a lot of behaviour that is deeply rooted in colonialism and patriarchy, yet we don't even realize it. So, it's important to understand this can happen, analyze what we need to change, and try to break free from these norms so that we can work towards decolonization.
-
-
www.darioamodei.com www.darioamodei.com
-
If AI further increases economic growth and quality of life in the developed world, while doing little to help the developing world, we should view that as a terrible moral failure
He calls out inequality as a real risk even in the "good" future. It shows that for him, success is not just technological, it's about whether the benefits reach everyone.
-
Fear is one kind of motivator, but it’s not enough: we need hope as well.
He's arguing that AI conversations shouldn't be all doom. People need something inspiring to work toward, not just warnings about what could go wrong.
-
At the same time, the vitality of democracy depends on harnessing new technologies to improve democratic institutions, not just responding to risks. A truly mature and successful implementation of AI has the potential to reduce bias and be fairer for everyone.
I don't see why this wouldn't be a capability based on my current understanding of AI. If the formation of the AI system and the data that it's built on lack bias, there is seemingly no reason for the system to later develop any new biases and either way that's something the system can be monitored for as a precaution.
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public review):
Summary:
This study advances the lab's growing body of evidence exploring higher-order learning and its neural mechanisms. They recently found that NMDA receptor activity in the perirhinal cortex was necessary for integrating stimulus-stimulus associations with stimulus-shock associations (mediated learning) to produce preconditioned fear, but it was not necessary for forming stimulus-shock associations. On the other hand, basolateral amygdala NMDA receptor activity is required for forming stimulus-shock memories. Based on these facts, the authors assessed: (1) why the perirhinal cortex is necessary for mediated learning but not direct fear learning, and (2) the determinants of perirhinal cortex versus basolateral amygdala necessity for forming direct versus indirect fear memories. The authors used standard sensory preconditioning and variants designed to manipulate the novelty and temporal relationship between stimuli and shock and, therefore, the attentional state under which associative information might be processed. Under experimental conditions where information would presumably be processed primarily in the periphery of attention (temporal distance between stimulus/shock or stimulus pre-exposure), perirhinal cortex NMDA receptor activation was required for learning indirect associations. On the other hand, when information would likely be processed in focal attention (novel stimulus contiguous with shock), basolateral amygdala NMDA activity was required for learning direct associations. Together, the findings indicate that the perirhinal cortex and basolateral amygdala subserve peripheral and focal attention, respectively. The authors provide support for their conclusions using careful, hypothesis-driven experimental design, rigorous methods, and integrating their findings with the relevant literature on learning theory, information processing, and neurobiology. Therefore, this work will be highly interesting to several fields.
Strengths:
(1) The experiments were carefully constructed and designed to test hypotheses that were rooted in the lab's previous work, in addition to established learning theory and information processing background literature.
(2) There are clear predictions and alternative outcomes. The provided table does an excellent job of condensing and enhancing the readability of a large amount of data.
(3) In a broad sense, attention states are a component of nearly every behavioral experiment. Therefore, identifying their engagement by dissociable brain areas and under different learning conditions is an important area of research.
(4) The authors clearly note where they replicated their own findings, report full statistical measures, effect sizes, and confidence intervals, indicating the level of scientific rigor.
(5) The findings raise questions for future experiments that will further test the authors' hypotheses; this is well discussed.
Weaknesses:
As a reader, it is difficult to interpret how first-order fear could be impaired while preconditioned fear is intact; it requires a bit of "reading between the lines".
We appreciate the Reviewer’s point and have attempted to address on lines 55-63 of the revised paper: “In a recent pair of studies, we extended these findings in two ways. First, we showed that S1 does not just form an association with shock in stage 2; it also mediates an association between S2 and the shock. Thus, S2 enters testing in stage 3 already conditioned, able to elicit fear responses (Wong et al., 2019). Second, we showed that this mediated S2-shock association requires NMDAR-activation in the PRh, as well as communication between the PRh and BLA (Wong et al., 2025). These findings raise two critical questions: 1) why is the PRh engaged for mediated conditioning of S2 but not for direct conditioning of S1; and 2) more generally, what determines whether the BLA and/or PRh is engaged for conditioning of the S1 and/or S2?”
Reviewer #2 (Public review):
Summary:
This paper continues the authors' research on the roles of the basolateral amygdala (BLA) and the perirhinal cortex (PRh) in sensory preconditioning (SPC) and second-order conditioning (SOC). In this manuscript, the authors explore how prior exposure to stimuli may influence which regions are necessary for conditioning to the second-order cue (S2). The authors perform a series of experiments which first confirm prior results shown by the author - that NMDA receptors in the PRh are necessary in SPC during conditioning of the first-order cue (S1) with shock to allow for freezing to S2 at test; and that NMDA receptors in the BLA are necessary for S1 conditioning during the S1-shock pairings. The authors then set out to test the hypothesis that the PRh encodes associations in a peripheral state of attention, whereas the BLA encodes associations in a focal state of attention, similar to the A1 and A2 states in Wagner's theory of SOP. To do this, they show that BLA is necessary for conditioning to S2 when the S2 is first exposed during a serial compound procedure - S2-S1-shock. To determine whether pre-exposure of S2 will shift S2 to a peripheral focal state, the authors run a design in which S2-S1 presentations are given prior to the serial compound phase. The authors show that this restores NMDA receptor activity within the PRh as necessary for the fear response to S2 at test. They then test whether the presence of S1 during the serial compound conditioning allows the PRh to support the fear responses to S2 by introducing a delay conditioning paradigm in which S1 is no longer present. The authors find that PRh is no longer required and suggest that this is due to S2 remaining in the primary focal state.
Strengths:
As with their earlier work, the authors have performed a rigorous series of experiments to better understand the roles of the BLA and PRh in the learning of first- and second-order stimuli. The experiments are well-designed and clearly presented, and the results show definitive differences in functionality between the PRh and BLA. The first experiment confirms earlier findings from the lab (and others), and the authors then build on their previous work to more deeply reveal how these regions differ in how they encode associations between stimuli. The authors have done a commendable job of pursuing these questions.
Table 1 is an excellent way to highlight the results and provide the reader with a quick look-up table of the findings.
Weaknesses:
The authors have attempted to resolve the question of the roles of the PRh and BLA in SPC and SOC, which the authors have explored in previous papers. Laudably, the authors have produced substantial results indicating how these two regions function in the learning of first- and second-order cues, providing an opportunity to narrow in on possible theories for their functionality. Yet the authors have framed this experiment in terms of an attentional framework and have argued that the results support this particular framework and hypothesis - that the PRh encodes peripheral and the BLA encodes focal states of learning. This certainly seems like a viable and exciting hypothesis, yet I don't see why the results have been completely framed and interpreted this way. It seems to me that there are still some alternative interpretations that are plausible and should be included in the paper.
We appreciate the Reviewer’s point and have attempted to address it on lines 566-594 of the Discussion: “An additional point to consider in relation to Experiments 3A, 3B, 4A and 4B is the level of surprise that rats experienced following presentations of the familiar S2 in stage 2. Specifically, in Experiments 3A and 3B, S2 was followed by the expected S1 (low surprise) and its conditioning required activation of NMDA receptors in the PRh and not the BLA. By contrast, in Experiments 4A and 4B, S2 was followed by omission of the expected S1 (high surprise) and its conditioning required activation of NMDA receptors in the BLA and not the PRh. This raises the possibility that surprise, or prediction error, also influences the way that S2 is processed in focal and peripheral states of attention. When prediction error is low, S2 is processed in the peripheral state of attention: hence, learning under these circumstances requires NMDA receptor activation in the PRh and not the BLA. By contrast, when prediction error is high, S2 is preserved in the focal state of attention: hence, learning under these circumstances requires NMDA receptor activation in the BLA and not the PRh. The impact of prediction error on the processing of S2 could be assessed using two types of designs. In the first design, rats are pre-exposed to S2-S1 pairings in stage 1 and this is followed by S2-S3-shock pairings in stage 2. The important feature of this design is that, in stage 2, the S2 is followed by surprise in omission of S1 and presentation of S3. Thus, if a large prediction error maintains processing of the familiar S2 in the BLA, we might expect that its conditioning in this design would require NMDA receptor activation in the BLA (in contrast to the results of Experiment 3B) and no longer require NMDA receptor activation in the PRh (in contrast to the results of Experiment 3A). In the second design, rats are pre-exposed to S2 alone in stage 1 and this is followed by S2-[trace]-shock pairings in stage 2. The important feature of this design is that, in stage 2, the S2 is not followed by the surprising omission of any stimulus. Thus, if a small prediction error shifts processing of the familiar S2 to the PRh, we might expect that its conditioning in this design would no longer require NMDA receptor activation in the BLA (in contrast to the results of Experiment 4B) but, instead, require NMDA receptor activation in the PRh (in contrast to the results of Experiment 4A). Future studies will use both designs to determine whether prediction error influences the processing of S2 in the focus versus periphery of attention and, thereby, whether learning about this stimulus requires NMDA receptor activation in the BLA or PRh.”
Reviewer #3 (Public review):
Summary:
This manuscript presents a series of experiments that further investigate the roles of the BLA and PRH in sensory preconditioning, with a particular focus on understanding their differential involvement in the association of S1 and S2 with shock.
Strengths:
The motivation for the study is clearly articulated, and the experimental designs are thoughtfully constructed. I especially appreciate the inclusion of Table 1, which makes the designs easy to follow. The results are clearly presented, and the statistical analyses are rigorous. My comments below mainly concern areas where the writing could be improved to help readers more easily grasp the logic behind the experiments.
Weaknesses:
(1) Lines 56-58: The two previous findings should be more clearly summarized. Specifically, it's unclear whether the "mediated S2-shock" association occurred during Stage 2 or Stage 3. I assume the authors mean Stage 2, but Stage 2 alone would not yet involve "fear of S2," making this expression a bit confusing.
We apologise for the confusion and have revised the summary of our previous findings on lines 55-63. The revised text now states: “In a recent pair of studies, we extended these findings in two ways. First, we showed that S1 does not just form an association with shock in stage 2; it also mediates an association between S2 and the shock. Thus, S2 enters testing in stage 3 already conditioned, able to elicit fear responses (Wong et al., 2019). Second, we showed that this mediated S2-shock association requires NMDAR-activation in the PRh, as well as communication between the PRh and BLA (Wong et al., 2025). These findings raise two critical questions: 1) why is the PRh engaged for mediated conditioning of S2 but not for direct conditioning of S1; and 2) more generally, what determines whether the BLA and/or PRh is engaged for conditioning of the S1 and/or S2?”
(2) Line 61: The phrase "Pavlovian fear conditioning" is ambiguous in this context. I assume it refers to S1-shock or S2-shock conditioning. If so, it would be clearer to state this explicitly.
Apologies for the ambiguity - we have omitted the term “Pavlovian” which may have been the source of confusion: The revised text on lines 60-63 now states: “These findings raise two critical questions: 1) why is the PRh engaged for mediated conditioning of S2 but not for direct conditioning of S1; and 2) more generally, what determines whether the BLA and/or PRh is engaged for conditioning of the S1 and/or S2?”
(3) Regarding the distinction between having or not having Stage 1 S2-S1 pairings, is "novel vs. familiar" the most accurate way to frame this? This terminology could be misleading, especially since one might wonder why S2 couldn't just be presented alone on Stage 1 if novelty is the critical factor. Would "outcome relevance" or "predictability" be more appropriate descriptors? If the authors choose to retain the "novel vs. familiar" framing, I suggest providing a clear explanation of this rationale before introducing the predictions around Line 118.
We have incorporated the suggestion regarding “predictability” while also retaining “novelty” as follows.
L76-85: “For example, different types of arrangements may influence the substrates of conditioning to S2 by influencing its novelty and/or its predictive value at the time of the shock, on the supposition that familiar stimuli are processed in the periphery of attention and, thereby, the PRh (Bogacz & Brown, 2003; Brown & Banks, 2015; Brown & Bashir, 2002; Martin et al., 2013; McClelland et al., 2014; Morillas et al., 2017; Murray & Wise, 2012; Robinson et al., 2010; Suzuki & Naya, 2014; Voss et al., 2009; Yang et al., 2023) whereas novel stimuli are processed in the focus of attention and, thereby, the amygdala (Holmes et al., 2018; Qureshi et al., 2023; Roozendaal et al., 2006; Rutishauser et al., 2006; Schomaker & Meeter, 2015; Wright et al., 2003).”
L116-120: “Subsequent experiments then used variations of this protocol to examine whether the engagement of NMDAR in the PRh or BLA for Pavlovian fear conditioning is influenced by the novelty/predictive value of the stimuli at the time of the shock (second implication of theory) as well as their distance or separation from the shock (third implication of theory; Table 1).”
(4) Line 121: This statement should refer to S1, not S2.
(5) Line 124: This one should refer to S2, not S1.
We have checked the text on these lines for errors and confirmed that the statements are correct. The lines encompassing this text (L121-130) are reproduced here for convenience:
(1) When rats are exposed to novel S2-S1-shock sequences, conditioning of S2 and S1 will be disrupted by a DAP5 infusion into the BLA but not into the PRh (Experiments 2A and 2B);
(2) When rats are exposed to S2-S1 pairings and then to S2-S1-shock sequences, conditioning of S2 will be disrupted by a DAP5 infusion into the PRh but not the BLA whereas conditioning of S1 will be disrupted by a DAP5 infusion into the BLA not the PRh (Experiments 3A and 3B);
(3) When rats are exposed to S2-S1 pairings and then to S2 (trace)-shock pairings, conditioning of S2 will be disrupted by a DAP5 into the BLA not the PRh (Experiments 4A and 4B).
(6) Additionally, the rationale for Experiment 4 is not introduced before the Results section. While it is understandable that Experiment 4 functions as a follow-up to Experiment 3, it would be helpful to briefly explain the reasoning behind its inclusion.
Experiment 4 follows from the results obtained in Experiment 3; and, as noted, the reasoning for its inclusion is provided locally in its introduction. We attempted to flag this experiment earlier in the general introduction to the paper; but this came at the cost of clarity to the overall story. As such, our revised paper retains the local introduction to this experiment. It is reproduced here for convenience:
“In Experiments 3A and 3B, conditioning of the pre-exposed S1 required NMDAR-activation in the BLA and not the PRh; whereas conditioning of the pre-exposed S2 required NMDAR-activation in the PRh and not the BLA. We attributed these findings to the fact that the pre-exposed S2 was separated from the shock by S1 during conditioning of the S2-S1-shock sequences in stage 2: hence, at the time of the shock, S2 was no longer processed in the focal state of attention supported by the BLA; instead, it was processed in the peripheral state of attention supported by the PRh.
“Experiments 4A and 4B employed a modification of the protocol used in Experiments 3A and 3B to examine whether a pre-exposed S1 influences the processing of a pre-exposed S2 across conditioning with S2-S1-shock sequences. The design of these experiments is shown in Figure 4A. Briefly, in each experiment, two groups of rats were exposed to a session of S2-S1 pairings in stage 1 and, 24 hours later, a session of S2-[trace]-shock pairings in stage 2, where the duration of the trace interval was equivalent to that of S1 in the preceding experiments. Immediately prior to the trace conditioning session in stage 2, one group in each experiment received an infusion of DAP5 or vehicle only into either the PRh (Experiment 4A) or BLA (Experiment 4B). Finally, all rats were tested with presentations of the S2 alone in stage 3. If the substrates of conditioning to S2 are determined only by the amount of time between presentations of this stimulus and foot shock in stage 2, the results obtained in Experiments 4A and 4B should be the same as those obtained in Experiments 3A and 3B: acquisition of freezing to S2 will require activation of NMDARs in the PRh and not the BLA. If, however, the presence of S1 in the preceding experiments (Experiments 3A and 3B) accelerated the rate at which processing of S2 transitioned from the focus of attention to its periphery, the results obtained in Experiments 4A and 4B will differ from those obtained in Experiments 3A and 3B. That is, in contrast to the preceding experiments where acquisition of freezing to S2 required NMDAR-activation in the PRh and not the BLA, here acquisition of freezing to S2 should require NMDAR-activation in the BLA but not the PRh.”
Reviewer #1 (Recommendations for the authors):
I greatly enjoyed reading and reviewing this manuscript, and so I only have boilerplate recommendations.
(1) I might add a couple of sentences discussing how/why preconditioned fear could be intact while first-order fear is impaired. Of course, if I am interpreting the provided interpretation correctly, the reason is that peripheral processing is still intact even when BLA NMDA receptors are blocked, and so mediated conditioning still occurs. Does this mean that mediated conditioning does not require learning the first-order relationship, and that they occur in parallel? Perhaps I just missed this, but I cannot help but wonder whether/how the psychological processes at play might change when first-order learning is impaired, so this would be greatly appreciated.
As noted above, we have revised the general introduction (around lines 55-59) to clarify that the direct S1-shock and mediated S2-shock associations form in parallel. Hence, manipulations that disrupt first-order fear to the S1 (such as a BLA infusion of the NMDA receptor antagonist, DAP5) do not automatically disrupt the expression of sensory preconditioned fear to the S2.
(2) Adding to the above - does the SOP or another theory predict serial vs parallel information flow from focal state to peripheral, or perhaps it is both to some extent?
SOP predicts both serial and parallel processing of information in its focal and peripheral states. That is, some proportion of the elements that comprise a stimulus may decay from the focal state of attention to the periphery (serial processing); hence, at any given moment, the elements that comprise a stimulus can be represented in both focal and peripheral states (parallel processing).
Given the nature of the designs and tools used in the present study (between-subject assessment of a DAP5 effect in the BLA or PRh), we selected parameters that would maximize the processing of the S2 and S1 stimuli in one or the other state of activation; hence the results of the present study. We are currently examining the joint processing of stimulus elements across focal and peripheral states using simultaneous recordings of activity in the BLA and PRh. These recordings are collected from rats trained in the different stages of a within-subject sensory preconditioning protocol. The present study created the basis for this work, which will be published separately in due course.
(3) The organization of PRh vs BLA is nice and consistent across each figure, but I would suggest adding any kind of additional demarcation beyond the colors and text, maybe just more space between AB / CD. The figure text indicating PRh/BLA is a bit small.
Thank you for the suggestion – we have added more space between the top and bottom panels of the figure.
(4) Line 496 typo ..."in the BLA but not the BLA".
Apologies for the type - this has been corrected.
Reviewer #2 (Recommendations for the authors):
I found the experiments to be extremely well-designed and the results convincing and exciting. The hypothesis of the focal and peripheral states of attention being encoded by BLA and PRh respectively, is enticing, yet as indicated in the public review, this does not seem to be the only possible interpretation. This is my only serious comment for the authors.
(1) I think it would be worth reframing the article slightly to give credence to alternative hypotheses. Not to say that the authors' intriguing hypothesis shouldn't be an integral part of the introduction, but no alternatives are mentioned. In experiment 2, could the fact that S2 is already being a predictor of S1, not block new learning to S2? In the framework of stimulus-stimulus associations, there would be no surprise in the serial-compound stage of conditioning at the onset of S1. This may prevent direct learning of the S2-shock association within the BLA. This type of association may as well (S2 predicts S1, but it's omitted), which could support learning by S2. fall under the peripheral/focal theory, but I don't think it's necessary to frame this possibility in terms of a peripheral/focal theory. To build on this alternative interpretation, the absence of S1 in experiment 4 may induce a prediction error. The peripheral and focal states appear to correspond to A2 and A1 in SOP extremely well, and I think it would potentially add interest and support. If the authors do intend to make the paper a strong argument for their hypothesis, perhaps a few additional experiments may be introduced. If the novelty of S2 is critical for S2 not to be processed in a focal state during the serial compound stage, could pre-exposure of S2 alone allow for dependence of S2-shock on the PRh? Assuming this is what the authors would predict, this might disentangle the S-S theory mentioned above from the peripheral/focal theory. Or perhaps run an experiment S2-X in stage 1 and S2-S1-shock in stage 2? This said, I think the experiments are more than sufficient for an exciting paper as is, and I don't think running additional experiments is necessary. I would only argue for this if the authors make a hard claim about the peripheral/focal theory, as is the case for the way the paper is currently written.
We appreciate the reviewer’s excellent point and suggestions. We have included an additional paragraph in the Discussion on page 24 (lines 566-594). “An additional point to consider in relation to Experiments 3A, 3B, 4A and 4B is the level of surprise that rats experienced following presentations of the familiar S2 in stage 2. Specifically, in Experiments 3A and 3B, S2 was followed by the expected S1 (low surprise) and its conditioning required activation of NMDA receptors in the PRh and not the BLA. By contrast, in Experiments 4A and 4B, S2 was followed by omission of the expected S1 (high surprise) and its conditioning required activation of NMDA receptors in the BLA and not the PRh. This raises the possibility that surprise, or prediction error, also influences the way that S2 is processed in focal and peripheral states of attention. When prediction error is low, S2 is processed in the peripheral state of attention: hence, learning under these circumstances requires NMDA receptor activation in the PRh and not the BLA. By contrast, when prediction error is high, S2 is preserved in the focal state of attention: hence, learning under these circumstances requires NMDA receptor activation in the BLA and not the PRh. The impact of prediction error on the processing of S2 could be assessed using two types of designs. In the first design, rats are pre-exposed to S2-S1 pairings in stage 1 and this is followed by S2-S3-shock pairings in stage 2. The important feature of this design is that, in stage 2, the S2 is followed by surprise in omission of S1 and presentation of S3. Thus, if a large prediction error maintains processing of the familiar S2 in the BLA, we might expect that its conditioning in this design would require NMDA receptor activation in the BLA (in contrast to the results of Experiment 3B) and no longer require NMDA receptor activation in the PRh (in contrast to the results of Experiment 3A). In the second design, rats are pre-exposed to S2 alone in stage 1 and this is followed by S2-[trace]-shock pairings in stage 2. The important feature of this design is that, in stage 2, the S2 is not followed by the surprising omission of any stimulus. Thus, if a small prediction error shifts processing of the familiar S2 to the PRh, we might expect that its conditioning in this design would no longer require NMDA receptor activation in the BLA (in contrast to the results of Experiment 4B) but, instead, require NMDA receptor activation in the PRh (in contrast to the results of Experiment 4A). Future studies will use both designs to determine whether prediction error influences the processing of S2 in the focus versus periphery of attention and, thereby, whether learning about this stimulus requires NMDA receptor activation in the BLA or PRh.”
(3) I was surprised the authors didn't frame their hypothesis more in terms of Wagner's SOP model. It was minimally mentioned in the introduction or the authors' theory if it were included more in the introduction. I was wondering whether the authors may have avoided this framing to avoid an expectation for modeling SOP in their design. If this were the case, I think the paper stands on its own without modeling, and at least for myself, a comparison to SOP would not require modeling of SOP. If this was the authors' concern for avoiding it, I would suggest to the authors that they need not be concerned about it.
We appreciate the endorsement of Wagner’s SOP theory as a nice way of framing our results. We are currently working on a paper in which we use simulations to show how Wagner’s theory can accommodate the present findings as well as others in the literature on sensory preconditioning. For this reason, we have not changed the current paper in relation to this point.
-
-
www.youtube.com www.youtube.com
-
we used a number of different proxies at 12 different sites, and they all recorded very clearly the effects of the great acceleration. And with that midpoint of about 1952.9 years, it all makes perfect sense. So it's not just the site at Crawford Lake, but all of the sites that we looked at showed a very very similar signal.
for - definition - anthropocene - synchronized signals of great acceleration at all 12 sites, not just Crawford Lake - Francine McCarthy, Brock University
-
-
thatvastvariety.substack.com thatvastvariety.substack.com
-
My Summary,
This article is all about Mastery. It's aimed at young 20 somethings but it just relevant to use here because we have not found a niche that resonates with our souls.
- Mastery is a process made up of multiple parts,
- Work in Public
- Get critical feedback
- Deliberate Practice
- Intelligent self reflection, see the questions in the article via my tags
- Social Networking to understand you niche, ask these people intelligent questions and create a knowledge garden
- Increase luck interface
-
-
download-files.wixmp.com download-files.wixmp.com
-
Afer painting this grim picture, they declared thatthe China that I was visiting, the China outside of those heavy doors that theyhad just eagerly denounced, was not in fact “the real China.”2 Te real China, aland of rites and etiquette (liyi zhi bang), and a global exemplar of morality andharmony, was based in the “Great Way” (da dao) that extended from the begin-ning of time to modernity.3 But this Great Way had been lost decades ago, andhad been replaced by an inferior way (xiao dao), in which people were solely con-cerned with convenience, ease, speed, money, and their own selfsh interests. Now,
This statement feels kind of wild, it’s like the teachers honestly believe they’re the ones preserving the “real” China. It shows how nostalgia can turn into a comforting fantasy people use to avoid facing how much the world has changed. It also makes me feel like the academy isn’t just teaching manners at all, it’s creating its own little imagined universe.
-
-
publish.obsidian.md publish.obsidian.md
-
Saladin and Richard are both remembered as brilliant generals, with Saladin being additionally remembered for his chivalry and Richard for his ferocious courage in battle and inspirational leadership of his troops.
It’s really interesting how their legacies balance each other out. Saladin is admired not just for his military skill but also for his sense of honor, while Richard is remembered for his raw bravery and ability to fire up his troops. Together, they make that era feel larger than life.
-
Theodoric imagined himself as the restorer and protector of "Roman" order in the west; Justinian saw him as just another barbarian.
It’s interesting that Theodoric saw himself as restoring Roman order while Justinian viewed him as a barbarian. I find it fascinating how perspective shaped their reputations. I find it also funny
-
-
www.reddit.com www.reddit.com
-
I speak from my experience on Olympia typewriters, and my engineers degree says the same: The most inner portion is not intended to recurve. Main springs wind in one direction, all the way. In this status, it's not usable. But fear not, you can rescue it. I rescued a main spring that had the inner connector broken off. Main springs (like all springs) tend to be very tough steel. You can simply break the faulty portion off. Then, you clamp the new end in grip pliers that double as a grip and a heat sink. A vice will do the same, but will be very finicky with the narrow curvature of the spring. Let that piece of the spring that you intend to bend into your new connector peek out of the pliers. Get a blow torch and heat that tiny piece to a bright red glow, then bend it one-third of the intended bend. Heat again, bend again, and once again. And now comes the most important part, the annealing: Take the spring out of the heat sink. Carefully apply heat to the new connector and the first tiny piece of the spring with your blow torch. You want to warm the connector, the bend and approximately 5mm or ¼ inch of the untouched spring. Don't allow it to glow. It may only make slight orange traces in the flame, but no more. Pull the flame away slowly, don't let it crash cool, don't blow on it, don't put it in water. Don't burn your fingers in the process. Your main spring is shorter now, but it will work just fine. Welcome to the world of typewriter blacksmiths.
https://www.reddit.com/r/TypewriterRepair/comments/1pbnomi/mainspring_twisted/
-
-
www.reddit.com www.reddit.com
-
Reply to u/banksclaud at https://reddit.com/r/typewriters/comments/1pf09vb/please_help_surprise_my_son/
Etsy can often have people flipping machines without having any work done, so be careful on what price you're paying for what you're getting. If it's over $350, it ought to fully serviced and have some sort of guarantee. Otherwise, find something at your local repair shop: https://site.xavier.edu/polt/typewriters/tw-repair.html
This question is asked so often, I've written up some good general advice which should apply to your child: https://boffosocko.com/2025/03/29/first-time-typewriter-purchases-with-specific-recommendations-for-writers/ For the age and your desire not to be bulky, go for a portable machine and not a larger standard or the more finnicky ultra-portables.
Some might opt for the brighter colored typewriters for kids for the "fun" factor, but I've found, having done a few type-ins with a huge variety of machines, that it's often the adults that are drawn to the colorful machines (which tend to be less well-built and plastic-y/cheaper) while kids will respond well to the older, duller vintage machines.
Here's a few 1950's advertisements directed at parents of kids just for fun: <br /> - https://www.youtube.com/watch?v=VTrkDa-GuSI<br /> - https://www.youtube.com/watch?v=bOIRul7pXDY
-
-
emergingethics.substack.com emergingethics.substack.com
-
This AI dependency also includes being less able to read by yourself; you might be anxious right now that this article isn’t an outline with catchy sub-headlines and emojis, or that it’s so long. While outlines and bullet points are faster to read and make it easier to see how the discussion flows, stripping down an article to its bare bones can flatten the discussion too much, causing nuance or details to be lost and moving too quickly to be digested. It’s the equivalent of wolfing down fast-food instead of savoring a memorable meal that was prepared with care and expertise.
This makes me reflect on how often I skim or rely on shortcuts instead of engaging with full texts. The paragraph also hints at a bigger concern: if students become too dependent on AI to simplify everything, they may lose the stamina and skill needed for close reading, critical thinking, and grappling with nuance. In this way, the issue isn’t just about AI use, it’s about how our reading habits and expectations are being reshaped, possibly at the cost of deeper comprehension.
-
-
www.archives.gov www.archives.gov
-
Today, education is perhaps the most important function of state and local governments.
The Court is reminding us that education isn’t just another public service but it’s the foundation for everything else in life. School is where kids learn how to navigate society, build skills, and prepare for adulthood. So if a child is denied a fair shot at school because of segregation, it’s not a small issue but it affects their entire future. That’s why the Court treats this as such a big constitutional problem.
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the original reviews
eLife Assessment
his valuable study presents a theoretical model of how punctuated mutations influence multistep adaptation, supported by empirical evidence from some TCGA cancer cohorts. This solid model is noteworthy for cancer researchers as it points to the case for possible punctuated evolution rather than gradual genomic change. However, the parametrization and systematic evaluation of the theoretical framework in the context of tumor evolution remain incomplete, and alternative explanations for the empirical observations are still plausible.
We thank the editor and the reviewers for their thorough engagement with our work. The reviewers’ comments have drawn our attention to several important points that we have addressed in the updated version. We believe that these modifications have substantially improved our paper.
There were two major themes in the reviewers’ suggestions for improvement. The first was that we should demonstrate more concretely how the results in the theoretical/stylized modelling parts of our paper quantitatively relate to dynamics in cancer.
To this end, we have now included a comprehensive quantification of the effect sizes of our results across large and biologically-relevant parameter ranges. Specifically, following reviewer 1’s suggestion to give more prominence to the branching process, we have added two figures (Fig S3-S4) quantifying the likelihood of multi-step adaptation in a branching process for a large range of mutation rates and birth-death ratios. Formulating our results in terms of birth-death ratios also allowed us to provide better intuition regarding how our results manifest in models with constant population size vs models of growing populations. In particular, the added figure (Fig S3) highlights that the effect size of temporal clustering on the probability of successful 2-step adaptation is very sensitive to the probability that the lineage of the first mutant would go extinct if it did not acquire a second mutation. As a result, the phenomenon we describe is biologically likely to be most effective in those phases during tumor evolution in which tumor growth is constrained. This important pattern had not been described sufficiently clearly in the initial version of our manuscript, and we thank both reviewers for their suggestions to make these improvements.
The second major theme in the reviewers’ suggestions was focused on how we relate our theoretical findings to readouts in genomic data, with both reviewers pointing to potential alternative explanations for the empirical patterns we describe.
We have now extended our empirical analyses following some of the reviewers’ suggestions. Specifically, we have included analyses investigating how the contribution of reactive oxygen species (ROS)-related mutation signatures correlates with our proxies for multi-step adaptation; and we have included robustness checks in which we use Spearman instead of Pearson correlations. Moreover, we have included more discussion on potential confounds and the assumptions going into our empirical analyses as well as the challenges in empirically identifying the phenomena we describe.
Below, we respond in detail to the individual comments made by each reviewer.
Public Reviews:
Reviewer #1 (Public review):
Summary:
Grasper et al. present a combined analysis of the role of temporal mutagenesis in cancer, which includes both theoretical investigation and empirical analysis of point mutations in TCGA cancer patient cohorts. They find that temporally elevated mutation rates contribute to cancer fitness by allowing fast adaptation when the fitness drops (due to previous deleterious mutations). This may be relevant in the case of tumor suppressor genes (TSG), which follow the 2-hit hypothesis (i.e., biallelic 2 mutations are necessary to deactivate TS), and in cases where temporal mutagenesis occurs (e.g., high APOBEC, ROS). They provide evidence that this scenario is likely to occur in patients with some cancer types. This is an interesting and potentially important result that merits the attention of the target audience. Nonetheless, I have some questions (detailed below) regarding the design of the study, the tools and parametrization of the theoretical analysis, and the empirical analysis, which I think, if addressed, would make the paper more solid and the conclusion more substantiated.
Strengths:
Combined theoretical investigation with empirical analysis of cancer patients.
Weaknesses:
Parametrization and systematic investigation of theoretical tools and their relevance to tumor evolution.
We sincerely thank Reviewer 1 for their comments. As communicated in more detail in the point-by-point replies to the “Recommendations for the authors”, we have revised the paper to address these comments in various ways. To summarize, Reviewer 1 asked for (1) more comprehensive analyses of the parameter space, especially in ranges of small fitness effects and low mutation rates; (2) additional clarifications on details of mechanisms described in the manuscript; and (3) suggested further robustness checks to our empirical analyses. We have addressed these points as follows: we have added detailed analyses of dynamics and effect sizes for branching processes (see Sections SI2 and SI3 in the Supplementary Information, as well as Figures S3 and S4). As suggested, these additions provide characterizations of effect sizes in biologically relevant parameter ranges (low mutation rates and smaller fitness effect sizes), and extend our descriptions to processes with dynamically changing population sizes. Moreover, we have added further clarifications at suggested points in the manuscript, e.g. to elaborate on the non-monotonicities in Fig 3. Lastly, we have undertaken robustness checks using Spearman rather than Pearson correlation coefficients to quantify relations between TSG deactivation and APOBEC signature contribution, and have performed analyses investigating dynamics of reactive oxygen species-associated mutagenesis instead of APOBEC.
Reviewer #2 (Public review):
This work presents theoretical results concerning the effect of punctuated mutation on multistep adaptation and empirical evidence for that effect in cancer. The empirical results seem to agree with the theoretical predictions. However, it is not clear how strong the effect should be on theoretical grounds, and there are other plausible explanations for the empirical observations.
Thank you very much for these comments. We have now substantially expanded our investigations of the parameter space as outlined in the response to the “eLife Assessment” above and in the detailed comments below (A(1)-A(3)) to convey more quantitative intuition for the magnitude of the effects we describe for different phases of tumor evolution. We agree that there could be potential additional confounders to our empirical investigations besides the challenges regarding quantification that we already described in our initial version of the manuscript. We have thus included further discussion of these in our manuscript (see replies to B(1)-B(3)), and we have expanded our empirical analyses as outlined in the response to the “eLife Assessment”.
For various reasons, the effect of punctuated mutation may be weaker than suggested by the theoretical and empirical analyses:
(A1) The effect of punctuated mutation is much stronger when the first mutation of a two-step adaptation is deleterious (Figure 2). For double inactivation of a TSG, the first mutation--inactivation of one copy--would be expected to be neutral or slightly advantageous. The simulations depicted in Figure 4, which are supposed to demonstrate the expected effect for TSGs, assume that the first mutation is quite deleterious. This assumption seems inappropriate for TSGs, and perhaps the other synergistic pairs considered, and exaggerates the expected effects.
Thank you for highlighting this discrepancy between Figure 2 and Figure 4. For computational efficiency and for illustration purposes, we had opted for high mutation rates and large fitness effects in Figure 2; however, our results are valid even in the setting of lower mutation rates and fitness effects. To improve the connection to Figure 4, and to address other related comments regarding parameter dependencies, we have now added more detailed quantification of the effects we describe (Figures SF3 and SF4) to the revised manuscript. These additions show that the effects illustrated in Figure 2 retain large effect sizes when going to much lower mutation rates and much smaller fitness effects. Indeed, while under high mutation rates we only see the large relative effects if the first mutation is highly deleterious, these large effects become more universal when going to low mutation rates.
In general, it is correct that the selective disadvantage (or advantage) conveyed by the first mutation affects the likelihood of successful 2-step adaptations. It is also correct that the magnitude of the ‘relative effect’
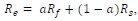 of temporal clustering on valley-crossing is highest if the lineage with only the first of the two mutations is vanishingly unlikely to produce a second mutant before going extinct. If the first mutation is strongly deleterious, the lineage of such a first mutant is likely to quickly go extinct – and therefore also more likely to do so before producing a second mutant.
of temporal clustering on valley-crossing is highest if the lineage with only the first of the two mutations is vanishingly unlikely to produce a second mutant before going extinct. If the first mutation is strongly deleterious, the lineage of such a first mutant is likely to quickly go extinct – and therefore also more likely to do so before producing a second mutant.However, this likelihood of producing the second mutant is also low if the mutation rate is low. As our added figure (Figure SF3) illustrates, at low mutation rates appropriate for cancer cells,
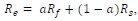 is insensitive to the magnitude of the fitness disadvantage for large parts of the parameter space. Especially in populations of constant size (approximated by a birth/death ratio of 1), the relative effects
is insensitive to the magnitude of the fitness disadvantage for large parts of the parameter space. Especially in populations of constant size (approximated by a birth/death ratio of 1), the relative effects 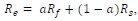 for first mutations that reduce the birth rate by 0.5 or by 0.05 are indistinguishable (Figure SF3f).
for first mutations that reduce the birth rate by 0.5 or by 0.05 are indistinguishable (Figure SF3f).Moreover, the absolute effect
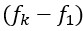 , as we discuss in the paper (Figures SF2 and SF3) is largest in regions of the parameter space in which the first mutant is not infinitesimally unlikely to produce a second mutant (and 𝑓<sub>𝑘</sub> and 𝑓<sub>1</sub> would be infinitesimally small), but rather in parameter regions in which this first mutant has a non-negligible chance to produce a second mutant. The absolute effect
, as we discuss in the paper (Figures SF2 and SF3) is largest in regions of the parameter space in which the first mutant is not infinitesimally unlikely to produce a second mutant (and 𝑓<sub>𝑘</sub> and 𝑓<sub>1</sub> would be infinitesimally small), but rather in parameter regions in which this first mutant has a non-negligible chance to produce a second mutant. The absolute effect 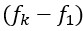 therefore peaks around fitness-neutral first mutations. While the next comment (below) says that our empirical investigations more closely resemble comparisons of relative effects and not absolute effects, we would expect that the observations in our data come preferentially from multi-step adaptations with large absolute effect since the absolute effect is maximal when both 𝑓<sub>𝑘</sub> and 𝑓<sub>1</sub>are relatively high.
therefore peaks around fitness-neutral first mutations. While the next comment (below) says that our empirical investigations more closely resemble comparisons of relative effects and not absolute effects, we would expect that the observations in our data come preferentially from multi-step adaptations with large absolute effect since the absolute effect is maximal when both 𝑓<sub>𝑘</sub> and 𝑓<sub>1</sub>are relatively high.In summary, we believe Figure 2, while having exaggerated parameters for very defendable reasons, is not a misleading illustration of the general phenomenon or of its applicability in biological settings, as effect sizes remain large when moving to biologically realistic parameter ranges. To clarify this issue, we have largely rewritten the relevant paragraphs in the results section and have added two additional figures (Figures SF3 and SF4) as well as a section in the SI with detailed discussion (SI2).
(A2) More generally, parameter values affect the magnitude of the effect. The authors note, for example, that the relative effect decreases with mutation rate. They suggest that the absolute effect, which increases, is more important, but the relative effect seems more relevant and is what is assessed empirically.
Thank you for this comment. As noted in the replies to the above comments, we have now included extensive investigations of how sensitive effect sizes are to different parameter choices. We also apologize for insufficiently clearly communicating how the quantities in Figure 4 relate to the findings of our theoretical models.
The challenge in relating our results to single-timepoint sequencing data is that we only observe the mutations that a tumor has acquired, but we do not directly observe the mutation rate histories that brought about these mutations. As an alternative readout, we therefore consider (through rough proxies: TSGs and APOBEC signatures) the amount of 2-step adaptations per acquired/retained mutation. While we unfortunately cannot control for the average mutation rate in a sample, we motivate using this “TSG-deactivation score” by the hypothesis that for any given mutation rate, we expect a positive relationship between the amount of temporal clustering and the amount of 2-step adaptations per acquired/retained mutation. This hypothesis follows directly from our theoretical model where it formally translates to the statement that for a fixed , is increasing in .
However, while both quantities 𝑓<sub>𝑘</sub>/𝑓<sub>1</sub> or
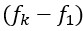 from our theoretical model relate to this hypothesis – both are increasing in 𝑘–, neither of them maps directly onto the formulation of our empirical hypothesis.
from our theoretical model relate to this hypothesis – both are increasing in 𝑘–, neither of them maps directly onto the formulation of our empirical hypothesis.We have now rewritten the relevant passages of the manuscript to more clearly convey our motivation for constructing our TSG deactivation score in this form (P. 4-6).
(A3) Routes to inactivation of both copies of a TSG that are not accelerated by punctuation will dilute any effects of punctuation. An example is a single somatic mutation followed by loss of heterozygosity. Such mechanisms are not included in the theoretical analysis nor assessed empirically. If, for example, 90% of double inactivations were the result of such mechanisms with a constant mutation rate, a factor of two effect of punctuated mutagenesis would increase the overall rate by only 10%. Consideration of the rate of apparent inactivation of just one TSG copy and of deletion of both copies would shed some light on the importance of this consideration.
This is a very good point, thank you. In our empirical analyses, the main motivation was to investigate whether we would observe patterns that are qualitatively consistent with our theoretical predictions, i.e. whether we would find positive associations between valley-crossing and temporal clustering. Our aim in the empirical analyses was not to provide a quantitative estimate of how strongly temporally clustered mutation processes affect mutation accumulation in human cancers. We hence restricted attention to only one mutation process which is well characterized to be temporally clustered (APOBEC mutagenesis) and to only one category of (epi)genomic changes (SNPs, in which APOBEC signatures are well characterized). Of course, such an analysis ignores that other mutation processes (e.g. LOH, copy number changes, methylation in promoter regions, etc.) may interact with the mechanisms that we consider in deactivating Tumor suppressor genes.
We have now updated the text to include further discussion of this limitation and further elaboration to convey that our empirical analyses are not intended as a complete quantification of the effect of temporal clustering on mutagenesis in-vivo (P. 10,11).
Several factors besides the effects of punctuated mutation might explain or contribute to the empirical observations:
(B1) High APOBEC3 activity can select for inactivation of TSGs (references in Butler and Banday 2023, PMID 36978147). This selective force is another plausible explanation for the empirical observations.
Thank you for making this point. We agree that increased APOBEC3 activity, or any other similar perturbation, can change the fitness effect that any further changes/perturbations to the cell would bring about. Our empirical analyses therefore rely on the assumption that there are no major confounding structural differences in selection pressures between tumors with different levels of APOBEC signature contributions. We have expanded our discussion section to elaborate on this potential limitation (P. 10-11).
While the hypothesis that APOBEC3 activity selects for inactivation of TSGSs has been suggested, there remain other explanations. Either way, the ways in which selective pressures have been suggested to change would not interfere relevantly with the effects we describe. The paper cited in the comment argues that “high APOBEC3 activity may generate a selective pressure favoring” TSG mutations as “APOBEC creates a high [mutation] burden, so cells with impaired DNA damage response (DDR) due to tumor suppressor mutations are more likely to avert apoptosis and continue proliferating”. To motivate this reasoning, in the same passage, the authors cite a high prevalence of TP53 mutations across several cancer types with “high burden of APOBEC3-induced mutations”, but also note that “this trend could arise from higher APOBEC3 expression in p53-mutated tumors since p53 may suppress APOBEC3B transcription via p21 and DREAM proteins”.
Translated to our theoretical framework, this reasoning builds on the idea that APOBEC3 activity increases the selective advantage of mutants with inactivation of both copies of a TSG. In contrast, the mechanism we describe acts by altering the chances of mutants with only one TSG allele inactivated to inactivate the second allele before going extinct. If homozygous inactivation of TSGs generally conveys relatively strong fitness advantages, lineages with homozygous inactivation would already be unlikely to go extinct. Further increasing the fitness advantage of such lineages would thus manifest mostly in a quicker spread of these lineages, rather than in changes in the chance that these lineages survive. In turn, such a change would have limited effect on the “rate” at which such 2-step adaptations occur, but would mostly affect the speed at which they fixate. It would be interesting to investigate these effects empirically by quantifying the speed of proliferation and chance of going extinct for lineages that newly acquired inactivating mutations in TSGs.
Beyond this explicit mention of selection pressures, the cited paper also discusses high occurrences of mutations in TSGs in relation to APOBEC. These enrichments, however, are not uniquely explained by an APOBEC-driven change in selection pressures. Indeed, our analyses would also predict such enrichments.
(B2) Without punctuation, the rate of multistep adaptation is expected to rise more than linearly with mutation rate. Thus, if APOBEC signatures are correlated with a high mutation rate due to the action of APOBEC, this alone could explain the correlation with TSG inactivation.
Thank you for making this point. Indeed, an identifying assumption that we make is that average mutation rates are balanced between samples with a higher vs lower APOBEC signature contribution. We cannot cleanly test this assumption, as we only observe aggregate mutation counts but not mutation rates. However, the fact that we observe an enrichment for APOBEC-associated mutations among the set of TSG-inactivating mutations (see Figure 4F) would be consistent with APOBEC-mutations driving the correlations in Fig 4D, rather than just average mutation rates. We have now added a paragraph to our manuscript to discuss these points (P. 10-11).
(B3) The nature of mutations caused by APOBEC might explain the results. Notably, one of the two APOBEC mutation signatures, SBS13, is particularly likely to produce nonsense mutations. The authors count both nonsense and missense mutations, but nonsense mutations are more likely to inactivate the gene, and hence to be selected.
Thank you for making this point. We have included it in our discussion of potential confounders/limitations in the revised manuscript (P. 10-11).
Recommendations for the authors:
Reviewer #1 (Recommendations for the authors):
Specific questions/comments/suggestions:
(1) For the theoretical investigation, the authors use the Wright-Fisher model with specific parameters for the decrease/increase in the fitness (0.5,1.5). This model is not so relevant to cancer, because it assumes a constant population size, while in cancer, the population is dynamic (increasing, if the tumor grows). Although I see they mention relevance to the branching process (in SI), I think the branching process should be bold in the main text and the Wright-Fisher in SI (or even dropped).
Thank you for this comment. We agree that too little attention had been given to the branching process in the original version of our manuscript. While the Wright-Fisher process is computationally efficient to simulate and thus lends itself to clean simulations for illustrative examples, it did lead us to put undue emphasis on populations of constant size.
The added Figures SF2 and SF3 now focus on branching processes, and we have substantially expanded our discussion of how dynamics differ as a function of the population-size trajectory (constant vs growing; SI2, P. 4,9,10). Generally, we do believe that it is appropriate to consider both regimes. If tumors evolve from being confined within their site of origin to progressively invading adjacent tissues and organ compartments, they traverse different regions of the birth-death ratio parameter space. Moreover, the timing of transitions between phases of more or less constrained growth is likely closely tied to adaptation dynamics, since breaching barriers to expansion requires adapting to novel environments and selection pressures.
We hope that the revised version of the manuscript conveys these points more clearly, and thank you for alerting us to this imbalance in the original version of our manuscript.
(2) The parameters 0.5 (decrease in fitness) and 1.5 (increase in fitness) seem exaggerated (the typical values for the selective advantage are usually much lower (by an order of magnitude). The same goes for the mutation rate. The authors chose values of the order 0.001, while in cancer (and generally) it is much lower than that (10-5 - 10-6). I think that generally, the authors should present a more systematic analysis of the sensitivity of the results to these parameters.
Thank you very much for this very important comment. We have made this a major focus in our revisions (see our reply to the editor’s comments). As suggested, we have now added further analyses to explore more biologically relevant parameter regimes. Reviewer 2 has made a similar remark, and to avoid redundancies, we point for a more detailed response to our response to that comment (A1).
(3) In Figure 3, the authors explore the sensitivity to mu (mutation rate) and k (temporal clustering) and find a non-monotonic behavior (Figure 3C). However, this behavior is not well explained. I think some more explanations are required here.
Thank you for pointing this out. We had initially relegated the more detailed explanations to the SI2 (which in the revised manuscript became SI4), but are happy to provide more elaboration in the main text, and have done so now (P. 5).
For , the non-monotonicity reflects the exploration-exploitation tradeoff that this section is dedicated to very small values (little exploration) prevent the population from finding fitness peaks. In contrast, once a fitness peak is reached, excessively large values (little exploitation) scatter the population away from this peak to points of lower fitness.
For , the most relevant dynamic is that at high , the population becomes unable to find close-by fitness improvements (1-step adaptations) if it is not in a burst. As 𝑘 increases, this delay in adaptation (until a burst occurs) eventually comes to outweigh the benefits of high 𝑘 (better ability to undergo multi-step adaptations). Additionally, if 𝑘 ∙ μ becomes very large, clonal interference eventually leads to diminishing exploration-returns when 𝑘 is increased further (Fig 5C), as the per-cell likelihood of finding a specific fitness peak eventually saturates and increasing only causes multiple cells to find the same peak, rather than one cell finding this peak and its lineage fixating in the population.
(4) In Figure 5, where the authors show the accumulation of the first (red; deleterious mutation) and second (blue; advantageous mutation), it seems that the fraction of deleterious mutations is much lower than that of advantageous mutations. This is opposite to the case of cancer, where most of the mutations are 'passengers', (slightly) deleterious or neutral mutations. Can the author explain this discrepancy and generally the relation of their parametrization to deleterious vs. advantageous mutations?
Thank you for this comment. In general, we have focused attention in our paper on sequences of mutations that bring about a fitness increase. We call those sequences ‘adaptations’ and categorize these as one-step or multi-step, depending on whether or not they contain intermediates states with a fitness disadvantage.
In our modelling, we do not consider mutations that are simply deleterious and are not a necessary part of a multi-step adaptation sequence. The motivation for this abstraction is, firstly, to focus on adaptation dynamics, and secondly, that in certain limits (small mu and large constant population sizes), lineages with only deleterious mutations have a probability close to one of going extinct, so that any emerging deleterious mutant would likely be 'washed out’ of the population before a new mutation emerges.
However, whether the dynamics of how neutral or deleterious passenger mutations are acquired also vary relevantly with the extent of temporal clustering is a valid and interesting question that would warrant its own study. The types of theoretical arguments for such an investigation would be very similar to the ones we use in our paper.
(5) The theoretical investigation assumes a multi/2-step adaptation scenario where the first mutation is deleterious and the second is advantageous. I think this should be generalized and further explored. For example, what happens when there are multiple mutations that are slightly deleterious (as probably is the case in cancer) and only much later mutations confer a selective advantage? How stable is the "valley crossing" if more deleterious mutations occur after the 2 steps?
This is also an important point and relates in part to the previous comment (4). For discussion of interactions with deleterious mutations, please see the reply to comment (4).
Regarding generalizations of this valley-crossing scenario, note that any sequence of mutations that increases fitness can be decomposed into sequences of either one-step or multi-step adaptations, as defined in the paper. Therefore, if all intermediate states before the final selectively advantageous state have a selective disadvantage making the lineages of such cells likely to go extinct, then our derivations in S1 apply, and the relative effect of temporal clustering becomes
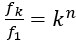 where n is the number of intermediate states. If, conversely, any of the intermediate states already had a selective advantage, then our model would consider the subsequence until this first mutation with a selective advantage as its individual (one-step or multi-step) “adaptation”.
where n is the number of intermediate states. If, conversely, any of the intermediate states already had a selective advantage, then our model would consider the subsequence until this first mutation with a selective advantage as its individual (one-step or multi-step) “adaptation”.The second question, “How stable is the "valley crossing" if more deleterious mutations occur after the 2 steps?”, touches on a different property of the population dynamics, namely on how the fate of a mutant lineage depends on how this lineage emerged. In our paper, we compare different levels of temporal clustering for a fixed average mutation rate. This choice implies that, if we assume that the mutant that emerges from a valley-crossing does not go extinct, then the number of deleterious mutations expected to occur in this lineage, once emerged, will not depend on the extent of temporal clustering. However, if in-burst mutation rates increased the expected burden of early acquired deleterious mutations sufficiently much to affect the probability that the lineage with a multi-step adaptation goes extinct before the burst ends, then there may indeed be an interaction between effects of deleterious passengers and temporal clustering. We would, however, expect effects on this probability of early extinction to be relatively minor, since such a lineage with a selective advantage would quickly grow to large cell-numbers implying that it would require a large number of co-occurring and sufficiently deleterious mutations across these cells for the lineage to go extinct.
(6) For the empirical analysis of TCGA cohorts, the authors focus on the contribution of APOBEC mutations (via signature analysis) to temporal mutagenesis. They find only a few cancer types (Figure 4D) that follow their prediction (in Figure 4C) of a correlation between TSG deactivation and temporal mutations in bursts. I think two main points should be addressed:
Thank you for this comment. We will respond in detail to the corresponding points below, but would like to note here that while we find this correlation “in only a few cancer types”, we also show that only few cancer types have relevant proportions of mutations caused by APOBEC, and it is precisely in these cancer types that we find a correlation. We have clarified this aspect in the revised version of the manuscript (P.7).
(i) APOBEC is not the only cause for temporal mutagenesis. For example, elevated ROS and hypoxia are also potential contributors - it might therefore be important to extend the signature analysis (to include more possible sources for temporal mutagenesis). Potentially, such an extension may show that more cancer types follow the author's prediction.
Thank you for this interesting suggestion. We have now included analogous analyses for contributions of signature SBS18 which is associated with ROS mutagenesis, and for the joint contribution of signatures SBS17a, SBS17b, SBS18 and SBS36, which all have been shown (some in a more context-dependent manner) to be associated with ROS mutagenesis. When doing so, we do not find a clear trend. However, we also do not find these signatures to account for substantial proportions of the acquired mutations, meaning that ROS mutagenesis likely also does not account for much of the variation in how temporally clustered the mutation rate trajectories of different tumors are. We have incorporated these results and their discussion in the manuscript (SI5 and Fig S8).
(ii) The TSG deactivation score used by the authors only counts the number of mutations and does not consider if the 2 mutations are biallelic, which is highly important in this case. There are ways to investigate the specific allele of mutations in TCGA data (for example, see Ciani et al. Cell Sys 2022 PMID: 34731645). Given the focus on TSG of this study, I think it is important to account for this in the analysis.
Thank you for making this point. We did initially consider inferring allele-specific mutation status, but decided against it as this would have shrunk our dataset substantially, thus potentially introducing unwanted biases. Determining whether two mutations lie on the same or on different alleles requires either (1) observing sequencing reads that either cover the loci of both mutations, or (2) tracing whether (sets of) other SNPs on the same gene co-occur exclusively with one of the two considered mutations. These requirements lead to a substantial filtering of the observed mutations. Moreover, this filtering would be especially strong for tumors with a small overall mutation burden, as these would have fewer co-occurring SNPs to leverage in this inference. We would have hence preferentially filtered out TSG-deactivating mutations in tumors with low mutation burden. We have modified the text to address this point (P.14).
(7) To continue point 4. I wonder why some known cancer types with high APOBEC signatures (e.g., lung, mentioned in the introduction) do not appear in the results of Figure 4. Can the author explain why it is missed?
We do provide complete results for all categories in Supplementary Figure 3. To not overwhelm the figure in the main text, we only show the four categories with the highest average APOBEC signature contribution, beyond those four, average APOBEC signature contributions quickly drop. Lung-related categories do not feature in these top four (Lung squamous cell carcinoma are fifth and Lung adenocarcinoma are eighth in this ordering).
Minors:
(1) It is worth mentioning the relevance to resistance to treatment (see https://www.nature.com/articles/s41588-025-02187-1).
Thank you for this suggestion. We have included a mention of the relation to this paper in the discussion section (P. 11).
(2) Some of the figures' resolution should be improved - specifically, Figures 4, S1, and S5, which are not clear/readable.
Thank you for pointing this out. This was the result of conversion to a word document. We will provide tif files in the revisions to have better resolution.
(3) Regarding Figure 3e,f. How come that moving from K=1 to K=I doesn't show any changes in fitness - it looks as if in both cases the value fluctuates around comparable mean fitness? Is that the case?
While fitness differences between simulations with different k manifest robustly over long time-horizons (see Fig 3C with results over generations), there are various sources of substantial stochasticity that make the fitness values in these short-term plots (Fig3D-F) imperfect illustrations of how long-term average fitness behaves. For instance, fitness landscapes are drawn randomly which introduces variability in how high and how close-by different fitness peaks are. Similarly, there is substantial randomness since both the type (direction on the 2-D fitness landscape) and the timing of mutation are stochastic.
The short-term plots in Fig3D-F are intended to showcase representative dynamics of transitions between points on the genotype space with different fitness values following a redrawing of the landscape – but not necessarily to provide a comparison between the height of the attained (local) fitness-maxima.
(4) Figures 4c,d - correlation should be Spearman, not Pearson (it's not a linear relationship).
Thank you for this comment. As a robustness check, we have generated the same figures using Spearman and not Pearson correlations and find results that are qualitatively consistent with the initially shown results. Indeed, using Spearman correlations, all four cancer types from Fig 4D have significant correlations.
(5) Typo for E) "...in samples of the cancer types in (C) were caused by APOBEC" - it should be D (not C) I guess.
Thank you for catching this. We fixed the typo.
(6) Figure 5 - the mutation rate is too high (0.001), sensitivity to that? Also the fitness change is exaggerated (0.5, 1.5), and the division of mutations to 100 and 100 (200 in total) loci is not clear.
Thank you for making this point. In this simulation setting it is unfortunately computationally prohibitively expensive to perform simulations at biologically realistic mutation rates. Therefore, we have scaled up the mutation rate while scaling down the population size. Moreover, the choice of model here is not meant to resemble a biologically realistic dynamic, but rather to create a stylized setting to be able to consider the interplay between clonal interference and facilitated valley-crossing in isolation. The key result from this figure is the separation of time scales at which low or high temporal clustering maximizes adaptability.
However, known parameter dependencies in these models allow us to reason about how tuning individual parameters of this stylized model would affect the relative importance of effects of clonal interference. This relative importance is largest when mutants are likely to co-occur on different competing clones in a population. The likelihood of such co-occurrences decreases substantially if decreasing the mutation rate to biologically realistic values. However, this likelihood also sensitively depends on the time that it takes a clone with a one-step adaptation to spread through the population. Smaller fitness advantages, as well as larger population sizes, slow down this process of taking over the population, which increases the likelihood of clonal interference. We now discuss these points in our revised manuscript (P. 8).
7) In the results text (last section) "Performing simulations for 2-step adaptations, we found that fixation rates are non-monotone in k. While at low k increasing k leads to a steep increase in the fixation rate, this trend eventually levels off and becomes negative, with further increases in k leading to a decrease in the fixation rate". Where are the results of this? It should be bold and apparent.
Thank you for alerting us that this is unclear. The relevant figure reference is indeed Fig 5C as in the preceding passage in the manuscript. However, we noticed that due to the presence of the steadily decreasing black line for 1-step adaptations, it is not easy to see that also the blue line is downward sloping. We have added a further reference to Fig 5C, and have adapted the grid spacing in the background of that figure-panel to make this trend more easily visible.
(8) Although not inconceivable, conclusions regarding resistance in the discussion are overstated. If you want to make this statement, you need to show that in resistant tumors, the temporal mutagenesis is responsible for progression vs. non-resistant/sensitive cases (is that the case), otherwise this should be toned down.
Thank you for pointing this out. We have tempered these conclusions in the revised version of the manuscript (P. 11).
Reviewer #2 (Recommendations for the authors):
(1) It might be useful to look specifically at X-linked TSGs. On the authors' interpretation, their relative inactivation rates should not be correlated with APOBEC signatures in males (but should be in females), though the size of the dataset may preclude any definite conclusions.
Thank you for this suggestion. Indeed, the size of the dataset unfortunately makes such analyses infeasible. Moreover, it is not clear whether X-linked TSGs might have structurally different fitness dynamics than TSGs on other chromosomes. However, this is an interesting suggestion worth following up on as more synergistic pairs confined to the X-chromosome are getting identified.
(2) Might there be value in distinguishing tumors that carry mutations expected to increase APOBEC expression from those that do not? Among several reasons, an APOBEC signature due to such a mutation and an APOBEC signature due to abortive viral infection may differ with respect to the degree of punctuation.
This is also an interesting suggestion for future investigations, but for which we unfortunately do not have sufficient information to build a meaningful analysis. In particular, it is unclear to what extent the degree and manifestation of episodicity/punctuation varies between these different mechanisms. Burst duration and intensity, as well as out-of-burst baseline rates of APOBEC mutagenesis likely differ in ways that are yet insufficiently characterized, which would make any result of analyses like these in Fig 4 hard to interpret.
(3) Also, in that paragraph, is "proportional to" used loosely to mean "an increasing function of"?
Thank you for this comment. We are not quite sure which paragraph is meant, but we use the term “proportional” in a literal sense at every point it is mentioned in the paper.
For the occurrences of the term on pages 3, 10 and 11, the word is used in reference to probabilities of reproduction (division in the branching process, or ‘being drawn to populate a spot in the next generation’ in the WF process) being “proportional” to fitness. These probabilities are constructed by dividing each individual cell’s fitness by the total fitness summed across all cells in the population. As the population acquires fitness-enhancing mutations, the resulting proportionality constant (1/total_fitness) changes, so that the mapping from ‘fitness’ to probability of reproduction in the next reproduction event changes over time. Nevertheless, this mapping always remains fitness-proportional.
On page 4, the term is used as follows: “the absolute rates 𝑓<sub>𝑘</sub> and 𝑓<sub>1</sub> are proportional to µ<sup>n+1”</sup>. Here, proportionality in the literal sense follows from the equations on page 20, when setting
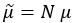 , so that the second factor becomes µ<sup>n+1</sup>. We have included a clarifying sentence to address this in the derivations (SI1).
, so that the second factor becomes µ<sup>n+1</sup>. We have included a clarifying sentence to address this in the derivations (SI1).(4) It could be mentioned in the main text that the time between bursts (d) must not be too short in order for the effect to be substantial. I would think that the relevant timescale depends on how deleterious the initial mutation is.
Thank you for making this interesting and very relevant point. We have included a section (SI3) and Figure (Fig S4) in the supplement to investigate the dependence on d. In short, we find that effects are weaker for small inter-burst intervals. The sensitivity to the burst size is highest for inter-burst intervals that are sufficiently small so that the lineage of the first mutant has relevant probability of surviving long enough to experience multiple burst phases.
(5) Why not report that relative rate for Figure 2E as for 2D, as the former would seem to be more relevant to TSGs? And why was it assumed that the first inactivation is deleterious in the simulations in Figure 4 if the goal is to model TSGs?
Thank you for noting this. For how we revised the paper to better connect Figures 2 and 4, please see our comment (A1) above. In general, neither 2E nor 2D should serve as quantitative predictions for what effect size we should expect in real world data, but are rather curated illustrations of the general phenomenon that we describe: we chose high mutation rates and exaggerated fitness effects so that dynamics become visually tractable in small simulation examples.
For figure 4, assuming that the first inactivation is deleterious achieves that the branching process for the mutant lineage becomes subcritical, which keeps the simulation example simple and illustrative. For more comprehensive motivation of the approach in 4D, and especially the discussion of how fitness effects of different magnitudes may or may not be subject to the effects we describe depending on whether the population is in a phase of constant or growing population size, we refer the reader to our added section SI2, and the added discussion on pages 6 and 10.
(6) Figure 2, D and E. I'm not sure why heatmaps with height one were provided rather than simple plots over time. It is difficult, for example, to determine from a heatmap whether the increase is linear or the relative rates with and without punctuation.
Thank you for this comment. These are not heatmaps with height one, but rather for every column of pixels, different segments of that column correspond to different clones within that population. This approach is intended to convey the difference in dynamics between the results in Fig 2 and the analogous results for a branching process in Fig S1. In Fig 2, valley-crossings happen sequentially, with subsequent fixations of adapted mutants. In Fig S1, with a growing population size, multiple clones with different numbers of adaptations coexist. We have now adapted the caption of Fig 2 to clarify this point.
(7) Page 3: "High mutation rates are known to limit the rate of 1-step adaptations due to clonal interference." This is a bit misleading, as it makes it sound like increasing the mutation rate decreases the rate of one-step adaptations.
Thank you for alerting us to this poor phrasing. We have changed it in the revised version of the manuscript (P. 3).
(8) Page 4: "proportional to \mu^{n+1}" Is "proportional" being used loosely for "an increasing function of"?
It is meant in the literal mathematical sense (see response to comment (3))
(9) Page 5, near bottom: "at least two mutations across the population". In the same genome?
We counted mutations irrespective of whether they emerged in the same genome, to remain analogous to the TCGA analyses for which we also do not have single cell-resolved information.
(10) Page 6: "missense or nonsense mutation". What about indels? If these are not affected by APOBEC, omitting them will exaggerate the effect of punctuation.
Thank you for pointing out that this focus on single nucleotide substitutions conveys an exaggerated image of the importance of this effect of APOBEC-driven mutagenesis. There are of course several other classes of (epi)genomic alterations (e.g. chromatin modifications, methylation changes, copy number changes) that we do not consider in this part of our analysis. APOBEC mutagenesis serves as an example of a temporally clustered mutation process, which we investigate in its domain of action.
We have added further discussion (P. 10-11) to convey that our empirical results merely constitute an investigation of whether empirical patterns are consistent with our hypothesis, but that the narrow focus on only SNVs, only TSGs, and only APOBEC mutagenesis does not allow for a general quantitative statement about the in-vivo relevance of the phenomena we describe.
(11) Page 6: "normalized by the total number of single nucleotide substitutions." It is difficult to know how to normalize correctly, but I might think that the square of the number of substitutions would be more appropriate. Perhaps the total numbers are close enough that it matters little.
Thank you for noting this. In the revised manuscript we have now expanded this passage in the text to more clearly convey our motivations for why we normalize by the total number of single nucleotide substitutions. While the likelihood for crossing a fitness valley with 2 mutations is indeed proportional to the square of the mutation rate, we do not directly observe mutation rates from our data. Rather, we observe the number of acquired single nucleotide substitutions for every tumor sample, but since tumors in our data differ in the time since initiation and therefore differ in the numbers of divisions their cells have undergone before being sequenced, we cannot directly infer mutation rates. One way to phrase our main result about valley-crossing is that temporally clustered mutation processes have an increased rate of successful valley-crossings per attempted valley crossing. Our TSG deactivation score is constructed to reflect this idea. The number of TSGs serves as a proxy for successful valley-crossings and the total mutation burden serves as a proxy for attempted valley-crossings.
To convey these points more clearly, we have rewritten the first paragraph in the Section “Proxies for valley crossing and for temporal clustering found in patient data” (P.6)
(12) Perhaps embed links to the COSMIC web pages for SBS2 and SBS13 in the text.
Thank you for this suggestion. We have embedded the links at the first mention of SBS2 and SBS13 in the text.
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public review):
In the Late Triassic and Early Jurassic (around 230 to 180 Ma ago), southern Wales and adjacent parts of England were a karst landscape. The caves and crevices accumulated remains of small vertebrates. These fossil-rich fissure fills are being exposed in limestone quarrying. In 2022 (reference 13 of the article), a partial articulated skeleton and numerous isolated bones from one fissure fill of end-Triassic age (just over 200 Ma) were named Cryptovaranoides microlanius and described as the oldest known squamate - the oldest known animal, by some 20 to 30 Ma, that is more closely related to snakes and some extant lizards than to other extant lizards. This would have considerable consequences for our understanding of the evolution of squamates and their closest relatives, especially for their speed and absolute timing, and was supported in the same paper by phylogenetic analyses based on different datasets.
In 2023, the present authors published a rebuttal (reference 18) to the 2022 paper, challenging anatomical interpretations and the irreproducible referral of some of the isolated bones to Cryptovaranoides. Modifying the datasets accordingly, they found Cryptovaranoides outside Squamata and presented evidence that it is far outside. In 2024 (reference 19), the original authors defended most of their original interpretation and presented some new data, some of it from newly referred isolated bones. The present article discusses anatomical features and the referral of isolated bones in more detail, documents some clear misinterpretations, argues against the widespread but not justifiable practice of referring isolated bones to the same species as long as there is merely no known evidence to the contrary, further argues against comparing newly recognized fossils to lists of diagnostic characters from the literature as opposed to performing phylogenetic analyses and interpreting the results, and finds Cryptovaranoides outside Squamata again.
Although a few of the character discussions and the discussion of at least one of the isolated bones can probably still be improved (and two characters are addressed twice), I see no sign that the discussion is going in circles or otherwise becoming unproductive. I can even imagine that the present contribution will end it.
We appreciate the positive response from reviewer 1!
Reviewer #2 (Public review):
Congratulations on this thorough manuscript on the phylogenetic affinities of Cryptovaranoides.
Thank you.
Recent interpretations of this taxon, and perhaps some others, have greatly changed the field's understanding of reptile origins- for better and (likely) for worse.
We agree, and note that while it is possible for challenges to be worse than the original interpretations, both the original and subsequent challenges are essential aspects of what make science, science.
This manuscript offers a careful review of the features used to place Cryptovaranoides within Squamata and adequately demonstrates that this interpretation is misguided, and therefore reconciles morphological and molecular data, which is an important contribution to the field of paleontology. The presence of any crown squamate in the Permian or Triassic should be met with skepticism, the same sort of skepticism provided in this manuscript.
We agree and add that every testable hypothesis requires skepticism and testing.
I have outlined some comments addressing some weaknesses that I believe will further elevate the scientific quality of the work. A brief, fresh read‑through to refine a few phrases, particularly where the discussion references Whiteside et al. could also give the paper an even more collegial tone.
We have followed Reviewer 2’s recommendations closely (see below) and have justified in our responses if we do not fully follow a particular recommendation.
This manuscript can be largely improved by additional discussion and figures, where applicable. When I first read this manuscript, I was a bit surprised at how little discussion there was concerning both non-lepidosauromorph lepidosaurs as well as stem-reptiles more broadly. This paper makes it extremely clear that Cryptovaranoides is not a squamate, but would greatly benefit in explaining why many of the characters either suggested by former studies to be squamate in nature or were optimized as such in phylogenetic analyses are rather widespread plesiomorphies present in crownward sauropsids such as millerettids, younginids, or tangasaurids. I suggest citing this work where applicable and building some of the discussion for a greatly improved manuscript. In sum:
(1) The discussion of stem-reptiles should be improved. Nearly all of the supposed squamate features in Cryptovaranoides are present in various stem-reptile groups. I've noted a few, but this would be a fairly quick addition to this work. If this manuscript incorporates this advice, I believe arguments regarding the affinities of Cryptovaranoides (at least within Squamata) will be finished, and this manuscript will be better off for it.
(2) I was also surprised at how little discussion there was here of putative stem-squamates or lepidosauromorphs more broadly. A few targeted comparisons could really benefit the manuscript. It is currently unclear as to why Cryptovaranoides could not be a stem-lepidosaur, although I know that the lepidosaur total-group in these manuscripts lacks character sampling due to their scarcity.
We are responding to (1) and (2) together. We agree with the Reviewer that a thorough comparison of Cryptovaranoides to non-lepidosaurian reptiles is critical. This is precisely what we did in our previous study: Brownstein et al. (2023)— see main text and supplementary information therein. As addressed therein, there is a substantial convergence between early lepidosaurs and some groups of archosauromorphs (our inferred position for Cryptovaranoides). Many of those points are not addressed in detail here in order to avoid redundancy and are simply referenced back to Brownstein et al. (2023). Secondly, stem reptiles (i.e., non-lepidosauromorphs and non-archosauromorphs), such as suggested above (millerettids, younginids, or tangasaurids), are substantially more distantly related to Cryptovaranoides (following any of the published hypotheses). As such, they share fewer traits (either symplesiomorphies or homoplasies), and so, in our opinion, we would risk directing losing the squamate-focus of our study.
We thus respectfully decline to engage the full scope of the problem in this contribution, but do note that this level of detailed work would make for an excellent student dissertation research program.
(3) This manuscript can be improved by additional figures, such as the slice data of the humerus. The poor quality of the scan data for Cryptovaranoides is stated during this paper several times, yet the scan data is often used as evidence for the presence or absence of often minute features without discussion, leaving doubts as to what condition is true. Otherwise, several sections can be rephrased to acknowledge uncertainty, and probably change some character scorings to '?' in other studies.
We strongly agree with the reviewer. Unfortunately, the original publication (Whiteside et al., 2021) did not make available the raw CT scan data to make this possible. As noted below in the Responses to Recommendations Section, we only have access to the mesh files for each segmented element. While one of us has observed the specimens personally, we have not had the opportunity to CT scan the specimens ourselves.
Reviewer #3 (Public review):
Summary:
The study provides an interesting contribution to our understanding of Cryptovaranoides relationships, which is a matter of intensive debate among researchers. My main concerns are in regard to the wording of some statements, but generally, the discussion and data are well prepared. I would recommend moderate revisions.
Strengths:
(1) Detailed analysis of the discussed characters.
(2) Illustrations of some comparative materials.
Thank you for noting the strengths inherent to our study.
Weaknesses:
Some parts of the manuscript require clarification and rewording.
One of the main points of criticism of Whiteside et al. is using characters for phylogenetic considerations that are not included in the phylogenetic analyses therein. The authors call it a "non-trivial substantive methodological flaw" (page 19, line 531). I would step down from such a statement for the reasons listed below:
(1) Comparative anatomy is not about making phylogenetic analyses. Comparative anatomy is about comparing different taxa in search of characters that are unique and characters that are shared between taxa. This creates an opportunity to assess the level of similarity between the taxa and create preliminary hypotheses about homology. Therefore, comparative anatomy can provide some phylogenetic inferences.
That does not mean that tests of congruence are not needed. Such comparisons are the first step that allows creating phylogenetic matrices for analysis, which is the next step of phylogenetic inference. That does not mean that all the papers with new morphological comparisons should end with a new or expanded phylogenetic matrix. Instead, such papers serve as a rationale for future papers that focus on building phylogenetic matrices.
We agree completely. We would also add that not every study presenting comparative anatomical work need be concluded with a phylogenetic analysis.
Our criticism of Whiteside et al. (2022) and (2024) is that these studies provided many unsubstantiated claims of having recovered synapomorphies between Cryptovaranoides and crown squamates without actually having done so through the standard empirical means (i.e., phylogenetic analysis and ancestral state reconstruction). Both Whiteside et al. (2022) and (2024) indicate characters presented as ‘shared with squamates’ along with 10 characters presented as synapomorphies (10). However, their actual phylogenetically recovered synapomorphies were few in number (only 3) and these were not discussed.
Furthermore, Whiteside et al. (2022) and (2024) comparative anatomy was restricted to comparing †Cryptovaranoides to crown squamates., based on the assumption that †Cryptovaranoides was a crown squamate and thus only needed to be compared to crown squamates.
In conclusion, we respectfully, we maintain such efforts are “non-trivial substantive methodological flaw(s)”.
(2) Phylogenetic matrices are never complete, both in terms of morphological disparity and taxonomic diversity. I don't know if it is even possible to have a complete one, but at least we can say that we are far from that. Criticising a work that did not include all the possibly relevant characters in the phylogenetic analysis is simply unfair. The authors should know that creating/expanding a phylogenetic matrix is a never-ending work, beyond the scope of any paper presenting a new fossil.
Respectfully, we did not criticize previous studies for including an incomplete phylogeny. Instead, we criticized the methodology behind the homology statements made in Whiteside et al. (2022) and Whiteside et al. (2024).
(3) Each additional taxon has the possibility of inducing a rethinking of characters. That includes new characters, new character states, character state reordering, etc. As I said above, it is usually beyond the scope of a paper with a new fossil to accommodate that into the phylogenetic matrix, as it requires not only scoring the newly described taxon but also many that are already scored. Since the digitalization of fossils is still rare, it requires a lot of collection visits that are costly in terms of time.
We agree on all points, but we are unsure of what the Reviewer is asking us to do relative to this study.
(4) If I were to search for a true flaw in the Whiteside et al. paper, I would check if there is a confirmation bias. The mentioned paper should not only search for characters that support Cryptovaranoides affinities with Anguimorpha but also characters that deny that. I am not sure if Whiteside et al. did such an exercise. Anyway, the test of congruence would not solve this issue because by adding only characters that support one hypothesis, we are biasing the results of such a test.
We would refer the Reviewer to their section (1) on comparative anatomy. As we and the Reviewer have pointed out, Whiteside et al. did not perform comparative anatomical statements outside of crown Squamata in their original study. More specifically, Whiteside et al. (2022, Fig. 8) presented a phylogeny where Cryptovaranoides formed a clade with Xenosaurus within the crown of Anguimorpha or what they termed “Anguiformes”, and made comparisons to the anatomies of the legless anguids, Pseudopus and Ophisaurus. Whiteside et al. (2024), abandoned “Anguiformes”, maintained comparisons to Pseudopus and emphasized affinities with Anguimorpha (but almost all of their phylogenies as published, they do not recover a monophyletic Angumimorpha unless amphisbaenians and snakes are considered to be anguimorphans. Thus, we agree that confirmation bias was inherent in their studies.
To sum up, there is nothing wrong with proposing some hypotheses about character homology between different taxa that can be tested in future papers that will include a test of congruence. Lack of such a test makes the whole argumentation weaker in Whiteside et al., but not unacceptable, as the manuscript might suggest. My advice is to step down from such strong statements like "methodological flaw" and "empirical problems" and replace them with "limitations", which I think better describes the situation.
We agree with the first sentence in this paragraph – there is nothing wrong with proposing character homologies between different taxa based on comparative anatomical studies. However, that is not what Whiteside et al. (2022) and (2024) did. Instead, they claimed that an ad hoc comparison of Cryptovaranoides to crown Squamata confirmed that Cryptovaranoides is in fact a crown squamate and likely a member of Anguimorpha. Their study did not recognize limitations, but rather, concluded that their new taxon pushed the age of crown Squamata into the Triassic.
As noted by Reviewer 2, such a claim, and the ‘data’ upon which it is based, should be treated with skepticism. We have elected to apply strong skepticism and stringent tests of falsification to our critique.
Reviewer #1 (Recommendations for the authors):
(1) Lines 596-598 promise the following: "we provide a long[-]form review of these and other features in Cryptovaranoides that compare favorably with non-squamate reptiles in Supplementary Material." You have kindly informed me that all this material has been moved into the main text; please amend this passage.
This has been deleted.
(2) Comments on science
41: I would rather say "an additional role".
This has been edited accordingly.
43: Reconstructing the tree entirely from extant organisms and adding fossils later is how Hennig imagined it, because he was an entomologist, and fossil insects are, on average,e extremely rare and usually very incomplete (showing a body outline and/or wing venation and little or nothing else). He was wrong, indeed wrong-headed. As a historical matter, phylogenetic hypotheses were routinely built on fossils by the mid-1860s, pretty much as soon as the paleontologists had finished reading On the Origin of Species, and this practice has never declined, let alone been interrupted. As a theoretical matter, including as many extinct taxa as possible in a phylogenetic analysis is desirable because it breaks up long branches (as most recently and dramatically shown by Mongiardino Koch & Parry 2020), and while some methods and some kinds of data are less susceptible to long-branch attraction and long-branch repulsion than others, none are immune; and while missing data (on average more common in fossils) can actively mislead parametric methods, this is not the case with parsimony, and even in Bayesian inference the problem is characters with missing data, not taxa with missing data. Some of you have, moreover, published tip-dated phylogenetic analyses. As a practical matter, molecular data are almost never available from fossils, so it is, of course, true that analyses which only use molecular data can almost never include fossils; but in the very rare exceptions, there is no reason to treat fossil evidence as an afterthought.
We agree and have changed “have become” to “is.”
49-50, 59: The ages of individual fissure fills can be determined by biostratigraphy; as far as I understand, all specimens ever referred to Cryptovaranoides [13, 19] come from a single fill that is "Rhaetian, probably late Rhaetian (equivalent of Cotham Member, Lilstock Formation)" [13: pp. 2, 15].
We appreciate this comment; the recent literature, however, suggests that variable ages are implied by the biostratigraphy at the English Fissure Fills, so we have chosen to keep this as is. Also note that several isolated bones were not recovered with the holotype but were discussed by Whiteside et al. (2024). The provenance of these bones was not clearly discussed in that paper.
59-60: Why "putative"? Just to express your disagreement? I would do that in a less misleading way, for example: "and found this taxon as a crown-group squamate (squamate hereafter) in their phylogenetic analyses." - plural because [19] presented four different analyses of two matrices just in the main paper.
We have removed this word.
121-124: The entepicondylar foramen is homologous all the way down the tree to Eusthenopteron and beyond. It has been lost a quite small number of times. The ectepicondylar foramen - i.e., the "supinator" (brachioradialis) process growing distally to meet the ectepicondyle, fusing with it and thereby enclosing the foramen - goes a bit beyond Neodiapsida and also occurs in a few other amniote clades (...as well as, funnily enough, Eusthenopteron in later ontogeny, but that's independent).
We agree. However, the important note here is that the features on the humerus of Cryptovaranoides are not comparable (differ in location and morphology) to the ent- and ectepondylar foramina in other reptiles, as we discuss at length. As such, we have kept this sentence as is.
153: Yes, but you [18] mistakenly wrote "strong anterior emargination of the maxillary nasal process, which is [...] a hallmark feature of archosauromorphs" in the main text (p. 14) - and you make the same mistake again here in lines 200-206! Also, the fact [19: Figure 2a-c] remains that Cryptovaranoides did not have an antorbital fenestra, let alone an antorbital fossa surrounding it (a fossa without a fenestra only occurs in some cases of secondary loss of the fenestra, e.g., in certain ornithischian dinosaurs). Unsurprisingly, therefore, Cryptovaranoides also does not have an orbital-as-opposed-to-nasal process on its maxilla [19: Figure 2a-c].
Line 243-249 (in original manuscript) deal with the emargination of maxillary nasal process (but this does not imply a full antorbital fenestra). We explicitly state that this feature alone "has limited utility" for supporting archosauromorph affinity.
158-173: The problem here is not that the capitellum is not preserved; from amniotes and "microsaurs" to lissamphibians and temnospondyls, capitella ossify late, and larger capitella attach to proportionately larger concave surfaces, so there is nothing wrong with "the cavity in which it sat clearly indicates a substantial condyle in life". Instead, the problem is a lack of quantification (...as has also been the case in the use of the exact same character in the debate on the origin of lissamphibians); your following sentence (lines 173-175) stands. The rest of the paragraph should be drastically shortened.
We appreciate this comment. We note that the ontogenetic variation of this feature is in part the issue with the interpretation provided by Whiteside et al. (2024). The issue is the lack of consistency on the morphology of the capitellum in that study. We are unclear on what the reviewer means by ‘quantification,’ as the character in question is binary.
250-252: It's not going to matter here, but in any different phylogenetic context, "sphenoid" would be confusing given the sphenethmoid, orbitosphenoid, pleurosphenoid, and laterosphenoid. I actually recommend "parabasisphenoid" as used in the literature on early amniotes (fusion of the dermal parasphenoid and the endochondral basisphenoid is standard for amniotes).
We have added "(=parabasisphenoid)" on first use but retain use of sphenoid because in the squamate and archosauromorph literature, sphenoid (or basisphenoid) is used more frequently.
314-315: Vomerine teeth are, of course, standard for sarcopterygians. Practically all extant amphibians have a vomerine toothrow, for example. A shagreen of denticles on the vomer is not as widespread but still reaches into the Devonian (Tulerpeton).
We agree, but vomerine teeth are rare in lepidosaurs and archosaurs and occur only in very recent clades e.g. anguids and one stem scincoid. Their presence in amphibians is not directly relevant to the phylogenetic placement of Cryptovaranoides among reptiles.
372: Fusion was not scored as present in [13], but as unknown (as "partial" uncertainty between states 0 and 1 [19:8]), and seemingly all three options were explored in [19].
We politely disagree with the reviewer; state 1 is scored in Whiteside et al. (2024).
377-383: Together with the partially fused NHMUK PV R37378 [13: Figure 4B, C; 19: 8], this is actually an argument that Cryptovaranoides is outside but close to Unidentata. The components of the astragalus fuse so early in extant amniotes that there is just a single ossification center in the already fused cartilage, but there are Carboniferous and Permian examples of astragali with sutures in the expected places; all of the animals in question (Diadectes, Hylonomus, captorhinids) seem to be close to but outside Amniota. (And yet, the astragalus has come undone in chamaeleons, indicating the components have not been lost.) - Also, if NHMUK PV R37378 doesn't belong to a squamate close to Unidentata, what does it belong to? Except in toothless beaks, premaxillary fusion is really rare; only molgin newts come to mind (and age, tooth size, and tooth number of NHMUK PV R37378 are wholly incompatible with a salamandrid).
The relevance of the astragalus is to the current discussion is unclear as we do not mention this element in our manuscript. We discuss the fusion in the premaxillae in response to previous comment.
471-474: That thing is concave. (The photo is good enough that you can enlarge it to 800% before it becomes too pixelated.) It could be a foramen filled with matrix; it does not look like a grain sticking to the outside of the bone. Also, spell out that you're talking about "suc.fo" in Figure 3j.
We are also a bit confused about this comment, as we state:
“Finally, we note here that Whiteside et al. [19] appear to have labeled a small piece of matrix attached to a coracoid that they refer to †C. microlanius as the supracoroacoid [sic] foramen in their figure 3, although this labeling is inferred because only “suc, supracoroacoid [sic]” is present in their figure 3 caption.” (L. 519-522, P. 17). We cannot verify that this structure is concave, as so we keep this text as is.
476-489: [19] conceded in their section 4.1 (pp. 11-12) that the atlas pleurocentrum, though fused to the dorsal surface of the axis intercentrum as usual for amniotes and diadectomorphs, was not fused to the axis pleurocentrum.
This is correct, as we note in the MS. The issue is whether these elements are clearly identifiable.
506-510: [19:12] did identify what they considered a possible ulnar patella, illustrated it (Figure 4d), scored it as unknown, and devoted the entire section 4.4 to it.<br /> 512-523: What I find most striking is that Whiteside et al., having just discovered a new taxon, feel so certain that this is the last one and any further material from that fissure must be referable to one of the species now known from there.
We agree with these points and believe we have devoted adequate text to addressing them. Note that the reviewer does not recommend any revisions to these sections.
553: Not that it matters, but I'm surprised you didn't use TNT 1.6; it came out in 2023 and is free like all earlier versions.
We have kept this as is following the reviewer comment, and because we were interested in replicating the analyses in the previous publications that have contributed to the debate about the identity of this taxon. For the present simple analyses both versions should perform identically, as the search algorithms for discrete characters are identical across these versions.
562: Is "01" a typo, or do you mean "0 or 1"? In that case, rather write "0/1" or "{01}".
This has been corrected to {01}
(3) Comments on nomenclature and terminology
55, 56: Delete both "...".
This has been corrected.
100: "ent- and ectepicondylar"
For clarity, we have kept the full words.
107-108: I understand that "high" is proximal and "low" is distal, but what is "the distal surface" if it is not the articular surface in the elbow joint?
This has been corrected.
120: "stem pan-lepidosaurs, and stem pan-squamates"; Lepidosauria and Squamata are crown groups that don't contain their stems
This has been corrected.
122, 123: Italics for Claudiosaurus and Delorhynchus.
This has been corrected.
130: Insert a space before "Tianyusaurus" (it's there in the original), and I recommend de-italicizing the two genus names to keep the contrast (as you did in line 162).
This has been corrected.
130, 131: Replace both "..." by "[...]", though you can just delete the second one.
This has been corrected.
174: Not a capitulum, but a grammatically even smaller (double diminutive) capitellum.
This has been corrected.
209, 224, Table 1: Both teams have consistently been doing this wrong. It's "recessus scalae tympani". The scala tympani ("ladder/staircase of the [ear]drum") isn't the recess, it's what the recess is for; therefore, the recess is named "recess of the scala tympani", and because there was no word for "of" in Classical Latin ("de" meant "off" and "about"), the genitive case was the only option. (For the same reason, the term contains "tympani", the genitive of "tympanum".)
This has been corrected.
415-425: This is a terminological nightmare. Ribs can have (and I'm not sure this is exhaustive): a) two separate processes (capitulum, tuberculum) that each bear an articulating facet, and a notch in between; b) the same, but with a non-articulating web of bone connecting the processes; c) a single uninterrupted elongate (even angled) articulating facet that articulates with the sutured or fused dia- and parapophysis; d) a single round articulating facet. Certainly, a) is bicapitate and d) is unicapitate, but for b) and c) all bets are off as to how any particular researcher is going to call them. This is a known source of chaos in phylogenetic analyses. I recommend writing a sentence or three on how the terms "unicapitate" & "bicapitate" lack fixed meanings and have caused confusion throughout tetrapod phylogenetics, and that the condition seen in Cryptovaranoides is nonetheless identical to that in archosauromorphs.
This has been added: “This confusion in part stems from the lack of a fixed meaning for uni- and bicapitate rib heads; in any case, †C. microlanius possesses a condition identical to archosauromorphs as we have shown.” (L.475-477, P.16).
439-440: Other than in archosaurs, some squamates and Mesosaurus, in which sauropsids are dorsal intercentra absent?
We are unclear about the relevance of the question to this section. The issue at hand is that some squamate lineages possess dorsal intercentra, so the absence of dorsal intercentra cannot be considered a squamate synapomorphy without the optimization of this feature along a phylogeny (which was not accomplished by Whiteside et al.).
458: prezygapophyses.
This has been corrected.
516: "[...]".
This has been corrected.
566: synapomorphies.
This has been corrected.
587: Macrocnemus.
This has been corrected.
585: I strongly recommend either taking off and nuking the name Reptilia from orbit (like Pisces) or using it the way it is defined in Phylonyms, namely as the crown group (a subset of Neodiapsida). Either would mean replacing "neodiapsid reptiles" with "neodiapsids".
This has been corrected to “neodiapsids.”
625: Replace "inclusive clades" by "included clades", "component clades", "subclades", or "parts," for example.
This has been kept as is because “inclusive clades” is common terminology and is used extensively in, for example, the PhyloCode.
659: Please update.
References are updated.
Fig. 8: Typo in Puercosuchus.
This has been corrected.
(4) Comments on style and spelling
You inconsistently use the past and the present tense to describe [13, 19], sometimes both in the same sentence (e.g., lines 323 vs. 325). I recommend speaking of published papers in the past tense to avoid ascribing past views and acts to people in their present state.
This has been corrected to be more consistent throughout the manuscript.
48: Remove the second comma.
This has been corrected.
91: Replace "[13] and WEA24" by "[13, 19]".
This has been corrected.
100: Commas on both sides of "in fact" or on neither
This has been corrected.
117: I recommend "the interpretation in [19]". I have nothing against the abbreviation "WEA24", but you haven't defined it, and it seems like a remnant of incomplete editing. - That said, eLife does not impose a format on such things. If you prefer, you can just bring citation by author & year back; in that case, this kind of abbreviation would make perfect sense (though it should still be explicitly defined).<br /> 129, 145: Likewise.
We have modified this [13] and [19] where necessary.
192-198: Surely this should be made part of the paragraph in lines 158-175, which has the exact same headline?
This has been corrected.
200-206: Surely this should be made part of the paragraph in lines 148-156, which has the exact same headline?
These sections deal with different issues pertaining to the analyses of Whiteside et al. (2024) and so we have kept to organization as is.
214: Delete "that".
This has been deleted.
312: "Vomer" isn't an adjective; I'd write "main vomer body" or "vomer's main body" or "main body of the vomer".
This has been corrected.
350: "figured"
This has been corrected.
400: Rather, "rearticulated" or "worked to rearticulate"? - And why "several"? Just write "two". "Several" implies larger numbers.
These issues have been corrected.
448, 500: As which? As what kind of feature? I'm aware that "as such" is fairly widely used for "therefore", but it still confuses me every time, and I have to suspect I'm not the only one. I recommend "therefore" or "for this reason" if that is what you mean.
“As such” has been deleted.
452: Adobe Reader doesn't let me check, but I think you have two spaces after "of".
This has been corrected.
514, 539, 546, 552, 588, Fig. 3, 5, 6, Table 1: "WEA24" strikes again.
This has been corrected.
515: Remove the parentheses.
This has been corrected.
531: Insert a space after the period.
This has been corrected.
532: Remove both commas and the second "that".
This has been corrected.
538: Remove the comma.
This has been kept as is because changing it would render the sentence grammatically incorrect.
545: "[...]" or, better, nothing.
This has been corrected.
547: Spaces on both sides of the dash or on neither (as in line 553).
This has been corrected.
552: Rather, "conducted a parsimony analysis".
This has been corrected.
556: Space after "[19]".
This has been corrected.
560: Comma after "narrow".
This has been corrected.
600: Comma after "above" to match the one in the preceding line - there's an insertion in the sentence that must be flanked by commas on both sides.
This has been corrected.
603: Compound adjectives like "alpha-taxonomic" need a hyphen to avoid tripping readers up.
This has been corrected.
612: Similarly, "ancestral-state reconstruction" needs one to make immediately clear it isn't a state reconstruction that is ancestral but a reconstruction of ancestral states.
This has been corrected.
613: If you want to keep this comma, you need to match it with another after "Cryptovaranoides" in line 611.
We have kept this as is, because removing this comma would render the sentence grammatically incorrect.
615: Likewise, you need a comma after "and" because "except for a few features" is an insertion. The other comma is actually optional; it depends on how much emphasis you want to place on what comes after it.
this has been added.
622: Comma after "[48, 49]".
this has been added.
672: Missing italics and two missing spaces.
This has been corrected.
678, 680-681, 693, 700-701, 734, 742, 747, 788, 797, 799, 803, 808, 810-811, 814, 817, 820, 823, 828, 841, 843: Missing italics.
This has been corrected.
683, 689: These are book chapters. Cite them accordingly.
This has been corrected.
737: Missing DOI.
No DOI is available.
793: Missing Bolosaurus major; and I'd rather cite it as "2024" than "in press", and "online early" instead of "n/a".
This has been corrected.
835: Hoffstetter, RJ?
This has been corrected.
836: Is there something missing?
This has been corrected.
839: This is the same reference as number 20 (lines 683-684), and it is miscited in a different way...!
This has been corrected.
Reviewer #2 (Recommendations for the authors):
(1) There is a brief mention of a phylogenetic analysis being re-run, but it is unclear if any modifications (changes in scoring) based on the very observations were made. Please state this explicitly.
This is explained from lines 600-622, P.20-21, in the section “Apomorphic characters not empirically obtained.” "In order to check the characters listed by Whiteside et al. [19] (p.19) as “two diagnostic characters” and “eight synapomorphies” in support of a squamate identity for †Cryptovaranoides, we conducted a parsimony analysis of the revised version of the dataset [32] provided by Whiteside et al. [19] in TNT v 1.5 [91]. We used Whiteside et al.’s [19] own data version"
(2) Line 20: There is almost no discussion of non‑lepidosaur lepidosauromorphs. I suggest including this, as the archosauromorph‑like features reported in Cryptovaranoides appear rather plastic. Furthermore, diagnostic features of Archosauromorpha in other datasets (e.g., Ezcurra 2016 or the works of Spiekman) are notably absent (and unsampled) in Cryptovaranoides. Expanding this comparison would greatly strengthen the manuscript.
The brief discussion (although not absent) of non-lepidosaur lepidosauromorphs is largely a function of the poor fossil record of this grade. But where necessary, we do discuss these taxa. Also see our previous study (Brownstein et al. 2023) for an extensive discussion of characters relevant to archosauromorphs.
(3) Line 38: I suggest removing "Archosauromorpha" from the keywords. The authors make a compelling case that Cryptovaranoides is not a squamate, yet they do not fully test its placement within Archosauromorpha (as they acknowledge). Perhaps use "Reptilia" instead?
We have removed this keyword.
(4) Line 99: The authors' points here are well made and largely valid. The presence of the ent‑ and ectepicondylar foramina is indeed an amniote plesiomorphy and cannot confirm a squamate identity. Their absence, however, can be informative - although it is unclear whether the CT scans of the humerus are of sufficient resolution, and Figure 4 of Brownstein et al. looks hastily reconstructed (perhaps owing to limited resolution). Moreover, the foramina illustrated by Whiteside do resemble those of other reptiles, albeit possibly over‑prepared and exaggerated.
The issue with the noted figure is indeed due to poor resolution from the scans. Although we agree with the reviewer, we hesitate to talk about absence in this taxon being phylogenetically informative given the confounding influence of ontogeny.
(5) I encourage the authors to provide slice data to support the claim that the foramina are absent (which could certainly be correct!); otherwise, the assertion remains unsubstantiated.
We only have access to the mesh files of segmented bones, not the raw (reconstructed slice) data.
(6) PLEASE NOTE - because the specimen is juvenile, the apparent absence of the ectepicondylar foramen is equivocal: the supinator process develops through ontogeny and encloses this foramen (see Buffa et al. 2025 on Thadeosaurus, for example).
See above.
(7) Line 122: Italicize 'Delorhynchus'
This has been corrected.
(8) Lines 131‑132: I'd suggest deleting the final sentence; it feels a little condescending, and your argument is already persuasive.
This has been corrected.
(9) Line 129: Please note that owenettid "parareptiles" also lack this process, as do several other stem‑saurians. Its absence is therefore not diagnostic of Squamata.<br /> Also: Such plasticity is common outside the crown. Milleropsis and Younginidae develop this process during ontogeny, even though a lower temporal bar never fully forms.
We appreciate this point. See discussion later in the manuscript.
(11) Line 172: Consider adding ontogeny alongside taphonomy and preservation. A juvenile would likely have a poorly developed radial condyle, if any. Acknowledging this possibility will add some needed nuance.
This sentence has been modified, but we have not added in discussion of ontogeny here because it is not immediately relevant to refuting the argument about inference of the presence of this feature when it is not preserved.
(12) Line 177: The "septomaxilla" in Whiteside et al. (2024, Figure 1C) resembles the contralateral premaxilla in dorsal view, with the maxillary process on the left and the palatal (or vomerine) process on the right (the dorsal process appears eroded). The foramen looks like a prepalatal foramen, common to many stem and crown reptiles. Consequently, scoring the septomaxilla as absent may be premature; this bone often ossifies late. In my experience with stem‑reptile aggregations, only one of several articulated individuals may ossify this element.
We agree that presence of a late-ossifying septomaxilla cannot be ruled out, but our point remains (and in agreement with Referee) that scoring the septomaxilla as present based on the amorphous fragments is premature.
(13) Line 200: Tomography data should be shown before citing it. The posterior margin of the maxilla appears rather straight, and the maxilla itself is tall for an archosauromorph. It would be more convincing to score this feature as present only after illustrating the relevant slices - and, as you note, the trait is widespread among non‑archosauromorphs.
See above and Brownstein et al. (2023).
(14) Line 208: Well argued: how could Whiteside et al. confidently assign a disarticulated element? Their "vagus" foramen actually resembles a standard hypoglossal foramen - identical to that seen in many stem reptiles, which often have one large and one small opening.
Thank you!
(15) Line 248: Again, please illustrate this region. One cannot argue for absence without showing the slice data. Note that millerettids and procolophonians - contemporaneous with Cryptovaranoides - possess an enclosed vidian canal, so the feature is broadly distributed.
See above.
(16) Line 258: The choanal fossa is intriguing: originally created for squamate matrices, yet present (to varying degrees) in nearly every reptile I have examined. It is strongly developed in millerettids (see Jenkins et al. 2025 on Milleropsis and Milleretta) and younginids, much like in squamates - Tiago appropriately scores it as present. Thus, it may be more of a "Neodiapsida + millerettids" character. In any case, the feature likely forms an ordered cline rather than a simple binary state.
We agree and look forward to future study of this feature.
(17) Line 283: Bolosaurids are not diapsids and, per Simões, myself, and others, "Diapsida" is probably invalid, at least how it is used here. Better to say "neodiapsids" for choristoderes and "stem‑reptiles" or "sauropsids" for bolosaurids. Jenkins et al.'s placement is largely a function of misidentifying the bolosaurid stapes as the opisthotic.
We are not entirely clear on this point since bolosaurids are not mentioned in this section.
(18) Line 298: Here, you note that the CT scans are rather coarse, which makes some earlier statements about absence/presence less certain (e.g., humeral foramina). It may strengthen the paper to make fewer definitive claims where resolution limits interpretation.
We appreciate this point. However, in the case of the humeral foramina the coarseness of the scans is one reason why we question Whiteside et al. scoring of the presence of these features.
(19) Line 314: Multiple rows of vomerine teeth are standard for amniotes; lepidosauromorphs such as Paliguana and Megachirella also exhibit them (though they may not have been segmented in the latter's description). Only a few groups (e.g., varanopids, some millerettids) have a single medial row.
We appreciate this point and have added in those citations into the following added sentence: “Multiple rows of vomerine teeth are common in reptiles outside of Squamata [76]; the presence of only one row is restricted to a handful of clades, including millerettids [77,78], †Tanystropheus [49], and some [79], but not all [71,80] choristoderes.” (L. 360-363, P. 12).
(20) Line 317: This is likely a reptile plesiomorphy - present in all millerettids (e.g., Milleropsis and Milleretta per Jenkins et al.). Citing these examples would clarify that it is not uniquely squamate. Could it be secondarily lost in archosauromorphs?
We appreciate this point and have cited Jenkins et al. here. It is out of the scope of this discussion to discuss the polarity of this feature relative to Archosauromorpha.
(21) Line 336: Unfortunately, a distinct quadratojugal facet is usually absent in Neodiapsids and millerettids; where present, the quadratojugal is reduced and simply overlaps the quadrate.
We appreciate this point but feel that reviewing the distribution of this feature across all reptiles is not relevant to the text noted.
(22) Line 357: Pterygoid‑quadrate overlap is likely a tetrapod plesiomorphy. Whiteside et al. do not define its functional or phylogenetic significance, and the overlap length is highly variable even among sister taxa.
We agree, but in any case this feature is impossible to assess in Cryptovaranoides.
(23) Line 365: Another well‑written section - clear and persuasive.
Thank you!
(24) Line 385: The cephalic condyle is widespread among neodiapsids, so it is not uniquely squamate.
We agree.
(25) Character 391: Note that the frontal underlapping the parietal is widespread, appearing in both millerettids and neodiapsids such as Youngina.
We appreciate this point, but the point here deals with the fact that this feature is not observable in the holotype of Cryptovaranoides.
(26) Line 415: The "anterior process" is actually common among crown reptiles, including sauropterygians, so it cannot by itself place Cryptovaranoides within Archosauromorpha.
We agree but also note that we do not claim this feature unambiguously unites Cryptovaranoides with Archosauromorpha.
(28) Line 460: Yes - Whiteside et al. appear to have relabeled the standard amniote coracoid foramen. Excellent discussion.
Thank you!
(29) Line 496: While mirroring Whiteside's structure, discussing this mandibular character earlier, before the postcrania, might aid readability.
We have chosen to keep this structure as is.
(30) Lines 486-588: This section oversimplifies the quadrate articulation.
We are unclear how this is an oversimplification.
(31) Both Prolacerta and Macrocnemus possess a cephalic condyle and some mobility (though less than many squamates). In Prolacerta (Miedema et al. 2020, Figure 4), the squamosal posteroventral process loosely overlaps the quadrate head.
We assume this comment refers to the section "Peg-in-notch articulation of quadrate head"; we appreciate clarification that this feature occurs in variable extent outside squamates, but this does not affect our statement that the material of Cryptovaranoides is too poorly preserved to confirm its presence.
(32) Where is this process in Cryptovaranoides? It is not evident in Whiteside's segmentation of the slender squamosal - please illustrate.
We are unclear as to which section this comment refers.
(33) Additionally, the quadrate "conch" of Cryptovaranoides is well developed, bearing lateral and medial tympanic crests; the lateral crest is absent in the cited archosauromorphs.
We note that no vertebrate has a medial tympanic crest (it is always laterally placed for the tympanic membrane, when present). If this is what the reviewer refers to, this is a feature commonly found across all tetrapods bearing a tympanum attached to the quadrate (e.g., most reptiles), and so it is not very relevant phylogenetically. Regarding its presence in Cryptovaranoides, the lateral margin of the quadrate is broken (Brownstein et al., 2023), so it cannot be determined. This incomplete preservation also makes an interpretation of a quadrate conch very hard to determine. But as currently preserved, there is no evidence whatsoever for this feature.
(34) Line 591: The cervical vertebrae of Cryptovaranoides are not archosauromorph‑like. Archosauromorph cervicals are elongate, parallelogram‑shaped, and carry long cervical ribs-none of which apply here. As the manuscript lacks a phylogenetic analysis, including these features seems unnecessary. Should they be added to other datasets, I suspect Cryptovaranoides would align along the lepidosaur stem (though that remains to be tested).
We politely disagree. The reviewer here mentions that the cervical vertebrae of archosauromorphs are generally shaped differently from those in Cryptovaranoides. The description provided (“elongate, parallelogram‑shaped, and carry long cervical ribs-none”) is basically limited to protorosaurians (e.g., tanystropheids, Macrocnemus) and early archosauriforms. We note that archosauromorph cervicals are notoriously variable in shape, especially in the crown, but also among early archosauromorphs. Further, the cervical ribs, are notoriously similar among early archosauromorphs (including protorosaurians) and Cryptovaranoides, as discussed and illustrated in Brownstein et al., 2023 (Figs. 2 and 3), especially concerning the presence of the anterior process.
Further, we do include a phylogenetic analysis of the matrix provided in Whiteside et al. (2024) as noted in our results section. In any case, we direct the reviewer to our previous study (Brownstein et al., 2023), in which we conduct phylogenetic analyses that included characters relevant to this note.
Reviewer #3 (Recommendations for the authors):
(1) The authors should use specimen numbers all over the text because we are talking about multiple individuals, and the authors contest the previous affinity of some of them. For example, on page 16, line 447, they mention an isolated vertebra but without any number. The specimen can be identified in the referenced article, but it would be much easier for the reader if the number were also provided here
Agreed and added.
(2) Abstract: "Our team questioned this identification and instead suggested Cryptovaranoides had unclear affinities to living reptiles."
That is very imprecise. The team suggested that it could be an archosauromorph or an indeterminate neodiapsid. Please change accordingly.
We politely disagree. We stated in our 2023 study that whereas our phylogenetic analyses place this taxon in Archosauromorpha, it remains unclear where it would belong within the latter. This is compatible with “unclear affinities to living reptiles”.
(3) Page 7, line 172: "Taphonomy and poor preservation cannot be used to infer the presence of an anatomical feature that is absent." Unfortunate wording. Taphonomy always has to be used to infer the presence or absence of anatomical features. Sometimes the feature is not preserved, but it leaves imprints/chemical traces or other taphonomic indicators that it was present in the organism. Please remove or rewrite the sentence.
We agree and have modified the sentence to read: “Taphonomy and poor preservation cannot be used alone to justify the inference that an anatomical feature was present when it is not preserved and there is no evidence of postmortem damage. In a situation when the absence of a feature is potentially ascribable to preservation, its presence should be considered ambiguous.” (L. 141-145, P.5).
(4) Page 4, line 91, please explain "WEA24" here, though it is unclear why this abbreviation is used instead of citation in the manuscript.
This has been corrected to Whiteside et al. [19].
(5) Page 6, line 144: "Together, these observations suggest that the presence of a jugal posterior process was incorrectly scored in the datasets used by WEA24 (type (ii) error)." That sentence is unclear. Why did the authors use "suggest"? Does it mean that they did not have access to the original data matrix to check it? If so, it should be clearly stated at the beginning of the manuscript.
See earlier; this has been modified and “suggest” has been removed.
(6) Page 7, line 174: "Finally, even in the case of the isolated humerus with a preserved capitulum, the condyle illustrated by Whiteside et al. [19] is fairly small compared to even the earliest known pan-squamates, such as Megachirella wachtleri (Figure 4)." Figure 4 does not show any humeri. Please correct.
The reference to figure 4 has been removed.
(7) Page 8, line 195-198: "This is not the condition specified in either of the morphological character sets that they cite [18,38], the presence of a distinct condyle that is expanded and is by their own description not homologous to the condition in other squamates." This is a bit unclear. Could the authors explain it a little bit further? How is the condition that is specified in the referred papers different compared to the Whiteside et al. description?
We appreciate this comment and have broken this sentence up into three sentences to clarify what we mean:
“The projection of the radial condyle above the adjacent region of the distal anterior extremity is not the condition specified in either of the morphological character sets that Whiteside et al. [19] cite [18,32]. The condition specified in those studies is the presence of a distinct condyle that is expanded. The feature described in Whiteside et al. [19] does not correspond to the character scored in the phylogenetic datasets.” (L.220-225, P.8).
(8) Page 16, line 446: "they observed in isolated vertebrae that they again refer to C. microlanius without justification". That is not true. The referred paper explains the attribution of these vertebrae to Cryptovaranoides (see section 5.3 therein). The authors do not have to agree with that justification, but they cannot claim that no justification was made. Please correct it here and throughout the text.
We have modified this sentence but note that the justification in Whiteside et al. (2024) lacked rigor. Whiteside et al. (2024) state: “Brownstein et al. [5] contested the affinities of three vertebrae, cervical vertebra NHMUK PV R37276, dorsal vertebra NHMUK PV R37277 and sacral vertebra NHMUK PV R37275. While all three are amphicoelous and not notochordal, the first two can be directly compared to the holotype. Cervical vertebra NHMUK PV R37276 is of the same form as the holotype CV3 with matching neural spine, ventral keel (=crest) and the posterior lateral ridges or lamina (figure 3c,d) shown by Brownstein et al. [5, fig. 1a]. The difference is that NHMUK PV R37276 has a fused neural arch to the pleurocentrum and a synapophysis rather than separate diapophysis and parapophysis of the juvenile holotype (figure 3c). Neurocentral fusion of the neural arch and centrum can occur late in modern squamates, ‘up to 82% of the species maximum size’ [28].
The dorsal surface of dorsal vertebra NHMUK PV R37277 (figure 3e) can be matched to the mid-dorsal vertebra in the †Cryptovaranoides holotype (figure 4d, dor.ve) and has the same morphology of wide, dorsally and outwardly directed, prezygapophyses, downwardly directed postzygapophyses and similar neural spine. It is also of similar proportions to the holotype when viewed dorsally (figures 3e and 4d), both being about 1.2 times longer anteroposteriorly than they are wide, measured across the posterior margin. The image in figure 4d demonstrates that the posterior vertebrae are part of the same spinal column as the truncated proximal region but the spinal column between the two parts is missing, probably lost in quarrying or fossil collection.”
This justification is based on pointing out the presence of supposed shared features between these isolated vertebrae and those in the holotype of Cryptovaranoides, even though none of these features are diagnostic for that taxon. We have changed the sentence in our manuscript to read:
“Whiteside et al. [19] concur with Brownstein et al. [18] that the diapophyses and parapophyses are unfused in the anterior dorsals of the holotype of †Cryptovaranoides microlanius, and restate that fusion of these structures is based on the condition they observed in isolated vertebrae that they refer to †C. microlanius based on general morphological similarity and without reference to diagnostic characters of †C. microlanius” (L. 502-507, P. 17).
(9) Figure 2. The figure caption lacks some explanations. Please provide information about affinity (e.g., squamate/gekkotan), ag,e and locality of the taxa presented. Are these left or right palatines? The second one seems to be incomplete, and maybe it is worth replacing it with something else?
The figure caption has been modified:
“Figure 2. Comparison of palatine morphologies. Blue shading indicates choanal fossa. Top image of †Cryptovaranoides referred left palatine is from Whiteside et al. [19]. Middle is the left palatine of †Helioscopos dickersonae (Squamata: Pan-Gekkota) from the Late Jurassic Morrison Formation [62]. Bottom is the right palatine of †Eoscincus ornatus (Squamata: Pan-Scincoidea) from the Late Jurassic Morrison Formation [31].”
(10) Figure 8. The abbreviations are not explained in the figure caption.
These have been added.
-
-
cutlefish.substack.com cutlefish.substack.com
-
This list has received positive feedback. It seems to have struck a nerve. Sending it to coincide with Monday morning to help folks navigate their week.Take care of yourself. Your brain is working overtime—all the time. Practice “radical” recovery.You may spend a lot longer thinking about things than most people. Pace your delivery.If you go deep first, and then simplify…keep in mind that you don’t need to show all of your work.Your default description of (almost) any problem will be too threatening/overwhelming.Do your deepest thinking with co-conspirators (not the people you’re trying to influence).Informal influence is often not formally recognized. Prepare mentally for this.The people you’re trying to influence spend 98% of their day overwhelmed by business as usual.Remember to also do the job you were hired to do (if you don’t you’ll be easier to discount).Seek “quick wins”, but know that most meaningful things will take a while.Some things take ages to materialize. It is discontinuous, not continuous.Make sure to celebrate your wins. They will be few and far between, so savor the moment.The people who support you in private may not be able to support you in public. Accept that.Hack existing power structures—it’s much easier than trying to change them.Consider becoming a formal leader. It’s harder in many ways, but you’ll have more leverage. What’s stopping you?In lieu of being a formal leader, make sure to partner with people who actually “own” the area of change.Watch out for imposing your worldview on people. Have you asked about what people care about?.You’ll need a support network. And not just a venting network. Real support.“Know when to fold ‘em”. Listen to Kenny Rogers The Gambler. Leave on your own terms.Don’t confuse being able to sense/see system dynamics, with being about to “control” them. You can’t.Grapple with your demons, and make sure not to wrap up too much of your identity in change.
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the previous reviews
Reviewer #1 (Public review):
Introduction & Theory
(1) It is difficult to appreciate why the first trial of extinction in a standard protocol does NOT produce the retrieval-extinction effect. This applies to the present study as well as others that have purported to show a retrieval-extinction effect. The importance of this point comes through at several places in the paper. E.g., the two groups in Study 1 experienced a different interval between the first and second CS extinction trials; and the results varied with this interval: a longer interval (10 min) ultimately resulted in less reinstatement of fear than a shorter interval. Even if the different pattern of results in these two groups was shown/known to imply two different processes, there is nothing in the present study that addresses what those processes might be. That is, while the authors talk about mechanisms of memory updating, there is little in the present study that permits any clear statement about mechanisms of memory. The references to a "short-term memory update" process do not help the reader to understand what is happening in the protocol.
We agree with the reviewer that whether and how the retrieval-extinction paradigm works is still under debate. Our results provide another line of evidence that such a paradigm is effective in producing long term fear amnesia. The focus of the current manuscript is to demonstrate that the retrieval-extinction paradigm can also facilitate a short-term fear memory deficit measured by SCR. Our TMS study provided some preliminary evidence in terms of the brain mechanisms involved in the causal relationship between the dorsolateral prefrontal cortex (dlPFC) activity and the short-term fear amnesia and showed that both the retrieval interval and the intact dlPFC activity were necessary for the short-term fear memory deficit and accordingly were referred to as the “mechanism” for memory update. We acknowledge that the term “mechanism” might have different connotations for different researchers. We now more explicitly clarify what we mean by “mechanisms” in the manuscript (line 99) as follows:
“In theory, different cognitive mechanisms underlying specific fear memory deficits, therefore, can be inferred based on the difference between memory deficits.”
In reply to this point, the authors cite evidence to suggest that "an isolated presentation of the CS+ seems to be important in preventing the return of fear expression." They then note the following: "It has also been suggested that only when the old memory and new experience (through extinction) can be inferred to have been generated from the same underlying latent cause, the old memory can be successfully modified (Gershman et al., 2017). On the other hand, if the new experiences are believed to be generated by a different latent cause, then the old memory is less likely to be subject to modification. Therefore, the way the 1stand 2ndCS are temporally organized (retrieval-extinction or standard extinction) might affect how the latent cause is inferred and lead to different levels of fear expression from a theoretical perspective." This merely begs the question: why might an isolated presentation of the CS+ result in the subsequent extinction experiences being allocated to the same memory state as the initial conditioning experiences? This is not yet addressed in any way.
As in our previous response, this manuscript is not about investigating the cognitive mechanism why and how an isolated presentation of the CS+ would suppress fear expression in the long term. As the reviewer is aware, and as we have addressed in our previous response letters, both the positive and negative evidence abounds as to whether the retrieval-extinction paradigm can successfully suppress the long-term fear expression. Previous research depicted mechanisms instigated by the single CS+ retrieval at the molecular, cellular, and systems levels, as well as through cognitive processes in humans. In the current manuscript, we simply set out to test that in addition to the long-term fear amnesia, whether the retrieval-extinction paradigm can also affect subjects’ short-term fear memory.
(2) The discussion of memory suppression is potentially interesting but, in its present form, raises more questions than it answers. That is, memory suppression is invoked to explain a particular pattern of results but I, as the reader, have no sense of why a fear memory would be better suppressed shortly after the retrieval-extinction protocol compared to the standard extinction protocol; and why this suppression is NOT specific to the cue that had been subjected to the retrieval-extinction protocol.
Memory suppression is the hypothesis we proposed that might be able to explain the results we obtained in the experiments. We discussed the possibility of memory suppression and listed the reasons why such a mechanism might be at work. As we mentioned in the manuscript, our findings are consistent with the memory suppression mechanism on at least two aspects: 1) cue-independence and 2) thought-control ability dependence. We agree that the questions raised by the reviewer are interesting but to answer these questions would require a series of further experiments to disentangle all the various variables and conceptual questions about the purpose of a phenomenon, which we are afraid is out of the scope of the current manuscript. We refer the reviewer to the discussion section where memory suppression might be the potential mechanism for the short-term amnesia we observed (lines 562-569) as follows:
“Previous studies indicate that a suppression mechanism can be characterized by three distinct features: first, the memory suppression effect tends to emerge early, usually 10-30 mins after memory suppression practice and can be transient (MacLeod and Macrae, 2001; Saunders and MacLeod, 2002); second, the memory suppression practice seems to directly act upon the unwanted memory itself (Levy and Anderson, 2002), such that the presentation of other cues originally associated with the unwanted memory also fails in memory recall (cue-independence); third, the magnitude of memory suppression effects is associated with individual difference in control abilities over intrusive thoughts (Küpper et al., 2014).”
(3) Relatedly, how does the retrieval-induced forgetting (which is referred to at various points throughout the paper) relate to the retrieval-extinction effect? The appeal to retrieval-induced forgetting as an apparent justification for aspects of the present study reinforces points 2 and 3 above. It is not uninteresting but lacks clarification/elaboration and, therefore, its relevance appears superficial at best.
We brought the topic of retrieval-induced forgetting (RIF) to stress the point that memory suppression can be unconscious. In a standard RIF paradigm, unlike the think/no-think paradigm, subjects are not explicitly told to suppress the non-target memories. However, to successfully retrieve the target memory, the cognitive system actively inhibits the non-target memories, effectively implementing a memory suppression mechanism (though unconsciously). Therefore, it is possible our results might be explained by the memory suppression framework. We elaborated this point in the discussion section (lines 578-584):
“In our experiments, subjects were not explicitly instructed to suppress their fear expression, yet the retrieval-extinction training significantly decreased short-term fear expression. These results are consistent with the short-term amnesia induced with the more explicit suppression intervention (Anderson et al., 1994; Kindt and Soeter, 2018; Speer et al., 2021; Wang et al., 2021; Wells and Davies, 1994). It is worth noting that although consciously repelling unwanted memory is a standard approach in memory suppression paradigm, it is possible that the engagement of the suppression mechanism can be unconscious.”
(4) I am glad that the authors have acknowledged the papers by Chalkia, van Oudenhove & Beckers (2020) and Chalkia et al (2020), which failed to replicate the effects of retrieval-extinction reported by Schiller et al in Reference 6. The authors have inserted the following text in the revised manuscript: "It should be noted that while our long-term amnesia results were consistent with the fear memory reconsolidation literature, there were also studies that failed to observe fear prevention (Chalkia, Schroyens, et al., 2020; Chalkia, Van Oudenhove, et al., 2020; Schroyens et al., 2023). Although the memory reconsolidation framework provides a viable explanation for the long-term amnesia, more evidence is required to validate the presence of reconsolidation, especially at the neurobiological level (Elsey et al., 2018). While it is beyond the scope of the current study to discuss the discrepancies between these studies, one possibility to reconcile these results concerns the procedure for the retrieval-extinction training. It has been shown that the eligibility for old memory to be updated is contingent on whether the old memory and new observations can be inferred to have been generated by the same latent cause (Gershman et al., 2017; Gershman and Niv, 2012). For example, prevention of the return of fear memory can be achieved through gradual extinction paradigm, which is thought to reduce the size of prediction errors to inhibit the formation of new latent causes (Gershman, Jones, et al., 2013). Therefore, the effectiveness of the retrieval-extinction paradigm might depend on the reliability of such paradigm in inferring the same underlying latent cause." Firstly, if it is beyond the scope of the present study to discuss the discrepancies between the present and past results, it is surely beyond the scope of the study to make any sort of reference to clinical implications!!!
As we have clearly stated in our manuscript that this paper was not about discussing why some literature was or was not able to replicate the retrieval-extinction results originally reported by Schiller et al. 2010. Instead, we aimed to report a novel short-term fear amnesia through the retrieval-extinction paradigm, above and beyond the long-term amnesia reported before. Speculating about clinical implications of these finding is unrelated to the long-term, amnesia debate in the reconsolidation world. We now refer the reader to several perspectives and reviews that have proposed ways to resolve these discrepancies as follows (lines 642-673).
Secondly, it is perfectly fine to state that "the effectiveness of the retrieval-extinction paradigm might depend on the reliability of such paradigm in inferring the same underlying latent cause..." This is not uninteresting, but it also isn't saying much. Minimally, I would expect some statement about factors that are likely to determine whether one is or isn't likely to see a retrieval-extinction effect, grounded in terms of this theory.
Again, as we have responded many times, we simply do not know why some studies were able to suppress the fear expression using the retrieval-extinction paradigm and other studies weren’t. This is still an unresolved issue that the field is actively engaging with, and we now refer the reader to several papers dealing with this issue. However, this is NOT the focus of our manuscript. Having a healthy debate does not mean that every study using the retrieval-extinction paradigm must address the long-standing question of why the retrieval-extinction paradigm is effective (at least in some studies).
Clarifications, Elaborations, Edits
(5) Some parts of the paper are not easy to follow. Here are a few examples (though there are others):
(a) In the abstract, the authors ask "whether memory retrieval facilitates update mechanisms other than memory reconsolidation"... but it is never made clear how memory retrieval could or should "facilitate" a memory update mechanism.
We meant to state that the retrieval-extinction paradigm might have effects on fear memory, above and beyond the purported memory reconsolidation effect. Sentence modified (lines 25-26) as follows:
“Memory reactivation renders consolidated memory fragile and thereby opens the window for memory updates, such as memory reconsolidation.”
(b) The authors state the following: "Furthermore, memory reactivation also triggers fear memory reconsolidation and produces cue specific amnesia at a longer and separable timescale (Study 2, N = 79 adults)." Importantly, in study 2, the retrieval-extinction protocol produced a cue-specific disruption in responding when testing occurred 24 hours after the end of extinction. This result is interesting but cannot be easily inferred from the statement that begins "Furthermore..." That is, the results should be described in terms of the combined effects of retrieval and extinction, not in terms of memory reactivation alone; and the statement about memory reconsolidation is unnecessary. One can simply state that the retrieval-extinction protocol produced a cue-specific disruption in responding when testing occurred 24 hours after the end of extinction.
The sentence the reviewer referred to was in our original manuscript submission but had since been modified based on the reviewer’s comments from last round of revision. Please see the abstract (lines 30-35) of our revised manuscript from last round of revision:
“Furthermore, across different timescales, the memory retrieval-extinction paradigm triggers distinct types of fear amnesia in terms of cue-specificity and cognitive control dependence, suggesting that the short-term fear amnesia might be caused by different mechanisms from the cue-specific amnesia at a longer and separable timescale (Study 2, N = 79 adults).”
(c) The authors also state that: "The temporal scale and cue-specificity results of the short-term fear amnesia are clearly dissociable from the amnesia related to memory reconsolidation, and suggest that memory retrieval and extinction training trigger distinct underlying memory update mechanisms." ***The pattern of results when testing occurred just minutes after the retrieval-extinction protocol was different to that obtained when testing occurred 24 hours after the protocol. Describing this in terms of temporal scale is unnecessary; and suggesting that memory retrieval and extinction trigger different memory update mechanisms is not obviously warranted. The results of interest are due to the combined effects of retrieval+extinction and there is no sense in which different memory update mechanisms should be identified with the different pattern of results obtained when testing occurred either 30 min or 24 hours after the retrieval-extinction protocol (at least, not the specific pattern of results obtained here).
Again, we are afraid that the reviewer referred to the abstract in the original manuscript submission, instead of the revised abstract we submitted in the last round. Please see lines 37-39 of the revised abstract where the sentence was already modified (or the abstract from last round of revision).
The facts that the 30min, 6hr and 24hr test results are different in terms of their cue-specificity and thought-control ability dependence are, to us, an important discovery in terms of delineating different cognitive processes at work following the retrieval-extinction paradigm. We want to emphasize that the fear memories after going through the retrieval-extinction paradigm showed interesting temporal dynamics in terms of their magnitudes, cue-specificity and thought-control ability dependence.
(d) The authors state that: "We hypothesize that the labile state triggered by the memory retrieval may facilitate different memory update mechanisms following extinction training, and these mechanisms can be further disentangled through the lens of temporal dynamics and cue-specificities." *** The first part of the sentence is confusing around usage of the term "facilitate"; and the second part of the sentence that references a "lens of temporal dynamics and cue-specificities" is mysterious. Indeed, as all rats received the same retrieval-extinction exposures in Study 2, it is not clear how or why any differences between the groups are attributed to "different memory update mechanisms following extinction"
The term “facilitate” was used to highlight the fact that the short-term fear amnesia effect is also memory retrieval dependent, as study 1 demonstrated. The novelty of the short-term fear memory deficit can be distinguished from the long-term memory effect via cue-specificity and thought-control ability dependence. Sentence has been modified (lines 97-101) as follows:
“We hypothesize that the labile state triggered by the memory retrieval may facilitate different memory deficits following extinction training, and these deficits can be further disentangled through the lens of temporal dynamics and cue-specificities. In theory, different cognitive mechanisms underlying specific fear memory deficits, therefore, can be inferred based on the difference between memory deficits.”
Data
(6A) The eight participants who were discontinued after Day 1 in Study 1 were all from the no reminder group. The authors should clarify how participants were allocated to the two groups in this experiment so that the reader can better understand why the distribution of non-responders was non-random (as it appears to be).
(6B) Similarly, in study 2, of the 37 participants that were discontinued after Day 2, 19 were from Group 30 min and 5 were from Group 6 hours. The authors should comment on how likely these numbers are to have been by chance alone. I presume that they reflect something about the way that participants were allocated to groups: e.g., the different groups of participants in studies 1 and 2 could have been run at quite different times (as opposed to concurrently). If this was done, why was it done? I can't see why the study should have been conducted in this fashion - this is for myriad reasons, including the authors' concerns re SCRs and their seasonal variations.
As we responded in the previous response letters (as well as in the revised the manuscript), subjects were excluded because their SCR did not reach the threshold of 0.02 S when electric shock was applied. Subjects were assigned to different treatments daily (eg. Day 1 for the reminder group and Day 2 for no-reminder group) to avoid potential confusion in switching protocols to different subjects within the same day. We suspect that the non-responders might be related to the body thermal conditions caused by the lack of central heating for specific dates. Please note that the discontinued subjects (non-responders) were let go immediately after the failure to detect their SCR (< 0.02 S) on Day 1 and never invited back on Day 2, so it’s possible that the discontinued subjects were all from certain dates on which the body thermal conditions were not ideal for SCR collection. Despite the number of excluded subjects, we verified the short-term fear amnesia effect in three separate studies, which to us should serve as strong evidence in terms of the validity of the effect.
(6C) In study 2, why is responding to the CS- so high on the first test trial in Group 30 min? Is the change in responding to the CS- from the last extinction trial to the first test trial different across the three groups in this study? Inspection of the figure suggests that it is higher in Group 30 min relative to Groups 6 hours and 24 hours. If this is confirmed by the analysis, it has implications for the fear recovery index which is partly based on responses to the CS-. If not for differences in the CS- responses, Groups 30 min and 6 hours are otherwise identical. That is, the claim of differential recovery to the CS1 and CS2 across time may simply an artefact of the way that the recovery index was calculated. This is unfortunate but also an important feature of the data given the way in which the fear recovery index was calculated.
We have provided detailed analysis to this question in our previous response letter, and we are posting our previous response there:
Following the reviewer’s comments, we went back and calculated the mean SCR difference of CS- between the first test trial and the last extinction trial for all three studies (see Author response image 1 below). In study 1, there was no difference in the mean CS- SCR (between the first test trial and last extinction trial) between the reminder and no-reminder groups (Kruskal-Wallis test
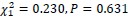 , though both groups showed significant fear recovery even in the CS- condition (Wilcoxon signed rank test, reminder: P = 0.0043, no-reminder: P = 0.0037). Next, we examined the mean SCR for CS- for the 30min, 6h and 24h groups in study 2 and found that there was indeed a group difference (one-way ANOVA,F<sub>2.76</sub> = 5.3462, P = 0.0067, panel b), suggesting that the CS- related SCR was influenced by the test time (30min, 6h or 24h). We also tested the CS- related SCR for the 4 groups in study 3 (where test was conducted 1 hour after the retrieval-extinction training) and found that across TMS stimulation types (PFC vs. VER) and reminder types (reminder vs. no-reminder) the ANOVA analysis did not yield main effect of TMS stimulation type (F<sub>1.71</sub> = 0.322, P = 0.572) nor main effect of reminder type (F<sub>1.71</sub> = 0.0499, P = 0.824, panel c). We added the R-VER group results in study 3 (see panel c) to panel b and plotted the CS- SCR difference across 4 different test time points and found that CS- SCR decreased as the test-extinction delay increased (Jonckheere-Terpstra test, P = 0.00028). These results suggest a natural “forgetting” tendency for CS- related SCR and highlight the importance of having the CS- as a control condition to which the CS+ related SCR was compared with.
, though both groups showed significant fear recovery even in the CS- condition (Wilcoxon signed rank test, reminder: P = 0.0043, no-reminder: P = 0.0037). Next, we examined the mean SCR for CS- for the 30min, 6h and 24h groups in study 2 and found that there was indeed a group difference (one-way ANOVA,F<sub>2.76</sub> = 5.3462, P = 0.0067, panel b), suggesting that the CS- related SCR was influenced by the test time (30min, 6h or 24h). We also tested the CS- related SCR for the 4 groups in study 3 (where test was conducted 1 hour after the retrieval-extinction training) and found that across TMS stimulation types (PFC vs. VER) and reminder types (reminder vs. no-reminder) the ANOVA analysis did not yield main effect of TMS stimulation type (F<sub>1.71</sub> = 0.322, P = 0.572) nor main effect of reminder type (F<sub>1.71</sub> = 0.0499, P = 0.824, panel c). We added the R-VER group results in study 3 (see panel c) to panel b and plotted the CS- SCR difference across 4 different test time points and found that CS- SCR decreased as the test-extinction delay increased (Jonckheere-Terpstra test, P = 0.00028). These results suggest a natural “forgetting” tendency for CS- related SCR and highlight the importance of having the CS- as a control condition to which the CS+ related SCR was compared with.Author response image 1.
(6D) The 6 hour group was clearly tested at a different time of day compared to the 30 min and 24 hour groups. This could have influenced the SCRs in this group and, thereby, contributed to the pattern of results obtained.
Again, we answered this question in our previous response. Please see the following for our previous response:
For the 30min and 24h groups, the test phase can be arranged in the morning, in the afternoon or at night. However, for the 6h group, the test phase was inevitably in the afternoon or at night since we wanted to exclude the potential influence of night sleep on the expression of fear memory (see Author response table 1 below). If we restricted the test time in the afternoon or at night for all three groups, then the timing of their extinction training was not matched.
Author response table 1.
Nevertheless, we also went back and examined the data for the subjects only tested in the afternoon or at nights in the 30min and 24h groups to match with the 6h group where all the subjects were tested either in the afternoon or at night. According to the table above, we have 17 subjects for the 30min group (9+8),18 subjects for the 24h group (9 + 9) and 26 subjects for the 6h group (12 + 14). As Author response image 2 shows, the SCR patterns in the fear acquisition, extinction and test phases were similar to the results presented in the original figure.
Author response image 2.
(6E) The authors find different patterns of responses to CS1 and CS2 when they were tested 30 min after extinction versus 24 h after extinction. On this basis, they infer distinct memory update mechanisms. However, I still can't quite see why the different patterns of responses at these two time points after extinction need to be taken to infer different memory update mechanisms. That is, the different patterns of responses at the two time points could be indicative of the same "memory update mechanism" in the sense that the retrieval-extinction procedure induces a short-term memory suppression that serves as the basis for the longer-term memory suppression (i.e., the reconsolidation effect). My pushback on this point is based on the notion of what constitutes a memory update mechanism; and is motivated by what I take to be a rather loose use of language/terminology in the reconsolidation literature and this paper specifically (for examples, see the title of the paper and line 2 of the abstract).
As we mentioned previously, the term “mechanism” might have different connotations for different researchers. We aim to report a novel memory deficit following the retrieval-extinction paradigm, which differed significantly from the purported reconsolidation related long-term fear amnesia in terms of its timescale, cue-specificity and thought-control ability. Further TMS study confirmed that the intact dlPFC function is necessary for the short-term memory deficit. It’s based on these results we proposed that the short-term fear amnesia might be related to a different cognitive “mechanism”. As mentioned above, we now clarify what we mean by “mechanism” in the abstract and introduction (lines 31-34, 97-101).
Reviewer #2 (Public review):
The fear acquisition data is converted to a differential fear SCR and this is what is analysed (early vs late). However, the figure shows the raw SCR values for CS+ and CS- and therefore it is unclear whether acquisition was successful (despite there being an "early" vs "late" effect - no descriptives are provided).
(1) There are still no descriptive statistics to substantiate learning in Experiment 1.
We answered this question in our previous response letter. We are sorry that the definition of “early” and “late” trials was scattered in the manuscript. For example, we wrote “the late phase of acquisition (last 5 trials)” (Line 375-376) in the results section. Since there were 10 trials in total for the acquisition stage, we define the first 5 trials and the last 5 trials as “early” and “late” phases of the acquisition stage and explicitly added them into the first occasion “early” and “late” terms appeared (lines 316-318).
In the results section, we did test whether the acquisition was successful in our previous manuscript (Line 316-325):
“To assess fear acquisition across groups (Figure 1B and C), we conducted a mixed two-way ANOVA of group (reminder vs. no-reminder) x time (early vs. late part of the acquisition; first 5 and last 5 trials, correspondingly) on the differential fear SCR. Our results showed a significant main effect of time (early vs. late; F<sub>1,55</sub> \= 6.545, P \= 0.013, η<sup>2</sup> \= 0.106), suggesting successful fear acquisition in both groups. There was no main effect of group (reminder vs. no-reminder) or the group x time interaction (group: F<sub>1,55</sub> \= 0.057, P \= 0.813, η<sup>2</sup> \= 0.001; interaction: F<sub>1,55</sub> \= 0.066, P \= 0.798, η<sup>2</sup> \= 0.001), indicating similar levels of fear acquisition between two groups. Post-hoc t-tests confirmed that the fear responses to the CS+ were significantly higher than that of CS- during the late part of acquisition phase in both groups (reminder group: t<sub>29</sub> \= 6.642, P < 0.001; no-reminder group: t<sub>26</sub> = 8.522, P < 0.001; Figure 1C). Importantly, the levels of acquisition were equivalent in both groups (early acquisition: t<sub>55</sub> \= -0.063, P \= 0.950; late acquisition: t<sub>55</sub> \= -0.318, P \= 0.751; Figure 1C).”
In Experiment 1 (Test results) it is unclear whether the main conclusion stems from a comparison of the test data relative to the last extinction trial ("we defined the fear recovery index as the SCR difference between the first test trial and the last extinction trial for a specific CS") or the difference relative to the CS- ("differential fear recovery index between CS+ and CS-"). It would help the reader assess the data if Fig 1e presents all the indexes (both CS+ and CS-). In addition, there is one sentence which I could not understand "there is no statistical difference between the differential fear recovery indexes between CS+ in the reminder and no reminder groups (P=0.048)". The p value suggests that there is a difference, yet it is not clear what is being compared here. Critically, any index taken as a difference relative to the CS- can indicate recovery of fear to the CS+ or absence of discrimination relative to the CS-, so ideally the authors would want to directly compare responses to the CS+ in the reminder and no-reminder groups. In the absence of such comparison, little can be concluded, in particular if SCR CS- data is different between groups. The latter issue is particularly relevant in Experiment 2, in which the CS- seems to vary between groups during the test and this can obscure the interpretation of the result.
(2) In the revised analyses, the authors now show that CS- changes in different groups (for example, Experiment 2) so this means that there is little to conclude from the differential scores because these depend on CS-. It is unclear whether the effects arise from CS+ performance or the differential which is subject to CS- variations.
There was a typo in the “P = 0.048” sentence and we have corrected it in our last response letter. Also in the previous response letter, we specifically addressed how the fear recovery index was defined (also in the revised manuscript).
In most of the fear conditioning studies, CS- trials were included as the baseline control. In turn, most of the analyses conducted also involved comparisons between different groups. Directly comparing CS+ trials across groups (or conditions) is rare. In our study 2, we showed that the CS- response decreased as a function of testing delays (30min, 1hr, 6hr and 24hr). Ideally, it would be nice to show that the CS- across groups/conditions did not change. However, even in those circumstances, comparisons are still based on the differential CS response (CS+ minus CS-), that is, the difference of difference. It is also important to note that difference score is important as CS+ alone or across conditions is difficult to interpret, especially in humans, due to noise, signal fluctuations, and irrelevant stimulus features; therefore trials-wise reference is essential to assess the CS+ in the context of a reference stimulus in each trial (after all, the baselines are different). We are listing a few influential papers in the field that the CS- responses were not particularly equivalent across groups/conditions and argue that this is a routine procedure (Kindt & Soeter 2018 Figs. 2-3; Sevenster et al., 2013 Fig. 3; Liu et al., 2014 Fig. 1; Raio et al., 2017 Fig. 2).
In experiment 1, the findings suggest that there is a benefit of retrieval followed by extinction in a short-term reinstatement test. In Experiment 2, the same effect is observed to a cue which did not undergo retrieval before extinction (CS2+), a result that is interpreted as resulting from cue-independence, rather than a failure to replicate in a within-subjects design the observations of Experiment 1 (between-subjects). Although retrieval-induced forgetting is cue-independent (the effect on items that are suppressed [Rp-] can be observed with an independent probe), it is not clear that the current findings are similar, and thus that the strong parallels made are not warranted. Here, both cues have been extinguished and therefore been equally exposed during the critical stage.
(3) The notion that suppression is automatic is speculative at best
We have responded the same question in our previous revision. Please note that our results from study 1 (the comparison between reminder and no-reminder groups) was not set up to test the cue-independence hypothesis for the short-term amnesia with only one CS+. Results from both study 2 (30min condition) and study 3 confirmed the cue-independence hypothesis and therefore we believe interpreting results from study 2 as “a failure to replicate in a within-subject design of the observations of Experiment 1” is not the case.
We agree that the proposal of automatic or unconscious memory suppression is speculative and that’s why we mentioned it in the discussion. The timescale, cue-specificity and the thought-control ability dependence of the short-term fear amnesia identified in our studies was reminiscent of the memory suppression effects reported in the previous literature. However, memory suppression typically adopted a conscious “suppression” treatment (such as the think/no-think paradigm), which was absent in the current study. However, the retrieval-induced forgetting (RIF), which is also considered a memory suppression paradigm via inhibitory control, does not require conscious effort to suppress any particular thought. Based on these results and extant literature, we raised the possibility of memory suppression as a potential mechanism. We make clear in the discussion that the suppression hypothesis and connections with RIF will require further evidence (lines 615-616):
“future research will be needed to investigate whether the short-term effect we observed is specifically related to associative memory or the spontaneous nature of suppression as in RIF (Figure 6C).”
(4) It still struggle with the parallels between these findings and the "limbo" literature. Here you manipulated the retention interval, whereas in the cited studies the number of extinction (exposure) was varied. These are two completely different phenomena.
We borrowed the “limbo” term to stress the transitioning from short-term to long-term memory deficits (the 6hr test group). Merlo et al. (2014) found that memory reconsolidation and extinction were dissociable processes depending on the extent of memory retrieval. They argued that there was a “limbo” transitional state, where neither the reconsolidation nor the extinction process was engaged. Our results suggest that at the test delay of 6hr, neither the short-term nor the long-term effect was present, signaling a “transitional” state after which the short-term memory deficit wanes and the long-term deficit starts to take over. We make this idea more explicit as follows (lines 622-626):
“These works identified important “boundary conditions” of memory retrieval in affecting the retention of the maladaptive emotional memories. In our study, however, we showed that even within a boundary condition previously thought to elicit memory reconsolidation, mnemonic processes other than reconsolidation could also be at work, and these processes jointly shape the persistence of fear memory.”
(5) My point about the data problematic for the reconsolidation (and consolidation) frameworks is that they observed memory in the absence of the brain substrates that are needed for memory to be observed. The answer did not address this. I do not understand how the latent cause model can explain this, if the only difference is the first ITI. Wouldn't participants fail to integrate extinction with acquisition with a longer ITI?
We take the sentence “they observed memory in the absence of the brain substrates that are needed for memory to be observed” as referring to the long-term memory deficit in our study. As we responded before, the aim of this manuscript was not about investigating the brain substrates involved in memory reconsolidation (or consolidation). Using a memory retrieval-extinction paradigm, we discovered a novel short-term memory effect, which differed from the purported reconsolidation effect in terms of timescale, cue-specificity and thought-control ability dependence. We further showed that both memory retrieval and intact dlPFC functions were necessary to observe the short-term memory deficit effect. Therefore, we conclude that the brain mechanism involved in such an effect should be different from the one related to the purported reconsolidation effect. We make this idea more explicit as follows (lines 546-547):
“Therefore, findings of the short-term fear amnesia suggest that the reconsolidation framework falls short to accommodate this more immediate effect (Figure 6A and B).”
Whilst I could access the data in the OFS site, I could not make sense of the Matlab files as there is no signposting indicating what data is being shown in the files. Thus, as it stands, there is no way of independently replicating the analyses reported.
(6) The materials in the OSF site are the same as before, they haven't been updated.
Last time we thought the main issue was the OSF site not being publicly accessible and thus made it open to all visitors. We have added descriptive file to explain the variables to help visitors to replicate the analyses we took.
(7) Concerning supplementary materials, the robustness tests are intended to prove that you 1) can get the same results by varying the statistical models or 2) you can get the same results when you include all participants. Here authors have done both so this does not help. Also, in the rebuttal letter, they stated "Please note we did not include non-learners in these analyses " which contradicts what is stated in the figure captions "(learners + non learners)"
In the supplementary materials, we did the analyses of varying the statistical models and including both learners and non-learners separately, instead of both. In fact, in the supplementary material Figs. 1 & 2, we included all the participants and performed similar analysis as in the main text and found similar results (learners + non-learners). Also, in the text of the supplementary material, we used a different statistical analysis method to only learners (analyzing subjects reported in the main text using a different method) and achieved similar results. We believe this is exactly what the reviewer suggested us to do. Also there seems to be a misunderstanding for the "Please note we did not include non-learners in these analyses" sentence in the rebuttal letter. As the reviewer can see, the full sentence read “Please note we did not include non-learners in these analyses (the texts of the supplementary materials)”. We meant to express that the Figures and texts in the supplementary material reflect two approaches: 1) Figures depicting re-analysis with all the included subjects (learners + non learners); 2) Text describing different analysis with learners. We added clarifications to emphasize these approaches in the supplementary materials.
(8) Finally, the literature suggesting that reconsolidation interference "eliminates" a memory is not substantiated by data nor in line with current theorising, so I invite a revision of these strong claims.
We agree and have toned down the strong claims.
Overall, I conclude that the revised manuscript did not address my main concerns.
In both rounds of responses, we tried our best to address the reviewer’s concerns. We hope that the clarifications in this letter and revisions in the text address the remaining concerns. Thank you for your feedback.
Reference:
Kindt, M. and Soeter, M. 2018. Pharmacologically induced amnesia for learned fear is time and sleep dependent. Nat Commun, 9, 1316.
Liu, J., Zhao, L., Xue, Y., Shi, J., Suo, L., Luo, Y., Chai, B., Yang, C., Fang, Q., Zhang, Y., Bao, Y., Pickens, C. L. and Lu, L. 2014. An unconditioned stimulus retrieval extinction procedure to prevent the return of fear memory. Biol Psychiatry, 76, 895-901.
Raio, C. M., Hartley, C. A., Orederu, T. A., Li, J. and Phelps, E. A. 2017. Stress attenuates the flexible updating of aversive value. Proc Natl Acad Sci U S A, 114, 11241-11246.
Sevenster, D., Beckers, T., & Kindt, M. 2013. Prediction error governs pharmacologically induced amnesia for learned fear. Science (New York, N.Y.), 339(6121), 830–833.
-
-
www.historians.org www.historians.org
-
History "offers the only extensive evidential base for the contemplation and analysis of how societies function, and people need to have some sense of how societies function simply to run their own lives."
Stearns is saying that history gives us the best information for understanding how societies work. If we don’t know how things worked in the past, it’s hard to understand how things work today. This matters for regular people too, not just historians, because it helps us make smarter decisions in everyday life.
-
-
utrgv-my.sharepoint.com utrgv-my.sharepoint.com
-
does multimodal mean?” Perhaps you remember an assignment from highschool when your teacher required you to create a Prezi or PowerPointpresentation, and she referred to it as a “multimodal project,”
his part helped me understand that multimodal composing isn’t just about adding pictures or videos for decoration. It’s about thinking intentionally about how different modes communicate meaning. I never really thought about writing like that before, like choosing tools based on what helps the reader.
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the original reviews
Public Reviews:
Reviewer #1 (Public review):
Summary:
Silbaugh, Koster, and Hansel investigated how the cerebellar climbing fiber (CF) signals influence neuronal activity and plasticity in mouse primary somatosensory (S1) cortex. They found that optogenetic activation of CFs in the cerebellum modulates responses of cortical neurons to whisker stimulation in a cell-type-specific manner and suppresses potentiation of layer 2/3 pyramidal neurons induced by repeated whisker stimulation. This suppression of plasticity by CF activation is mediated through modulation of VIP- and SST-positive interneurons. Using transsynaptic tracing and chemogenetic approaches, the authors identified a pathway from the cerebellum through the zona incerta and the thalamic posterior medial (POm) nucleus to the S1 cortex, which underlies this functional modulation.
Strengths:
This study employed a combination of modern neuroscientific techniques, including two-photon imaging, opto- and chemo-genetic approaches, and transsynaptic tracing. The experiments were thoroughly conducted, and the results were clearly and systematically described. The interplay between the cerebellum and other brain regions - and its functional implications - is one of the major topics in this field. This study provides solid evidence for an instructive role of the cerebellum in experience-dependent plasticity in the S1 cortex.
Weaknesses:
There may be some methodological limitations, and the physiological relevance of the CFinduced plasticity modulation in the S1 cortex remains unclear. In particular, it has not been elucidated how CF activity influences the firing patterns of downstream neurons along the pathway to the S1 cortex during stimulation.
Our study addresses the important question of whether CF signaling can influence the activity and plasticity of neurons outside the olivocerebellar system, and further identifies the mechanism through which this indeed occurs. We provide a detailed description of the involvement of specific neuron subtypes and how they are modulated by climbing fiber activation to impact S1 plasticity. We also identify at least one critical pathway from the cerebellar output to the S1 circuit. It is indeed correct that we did not investigate how the specific firing patterns of all of these downstream neurons are affected, or the natural behaviors in which this mechanism is involved. Now that it is established that CF signaling can impact activity and plasticity outside the olivocerebellar system -- and even in the primary somatosensory cortex -- these questions will be important to further investigate in future studies.
(1) Optogenetic stimulation may have activated a large population of CFs synchronously, potentially leading to strong suppression followed by massive activation in numerous cerebellar nuclear (CN) neurons. Given that there is no quantitative estimation of the stimulated area or number of activated CFs, observed effects are difficult to interpret directly. The authors should at least provide the basic stimulation parameters (coordinates of stim location, power density, spot size, estimated number of Purkinje cells included, etc.).
As discussed in the paper, we indeed expect that synchronous CF activation is needed to allow for an effect on S1 circuits under natural or optogenetic activation conditions. The basic optogenetic stimulation parameters (also stated in the methods) are as follows: 470 nm LED; Ø200 µm core, 0.39 NA rotary joint patch cable; absolute power output of 2.5 mW; spot size at the surface of the cortex 0.6 mm; estimated power density 8 mW/mm2. A serious estimate of the number of Purkinje cells that are activated is difficult to provide, in particular as ‘activation’ would refer to climbing fiber inputs, not Purkinje cells directly.
(2) There are CF collaterals directly innervating CN (PMID:10982464). Therefore, antidromic spikes induced by optogenetic stimulation may directly activate CN neurons. On the other hand, a previous study reported that CN neurons exhibit only weak responses to CF collateral inputs (PMID: 27047344). The authors should discuss these possibilities and the potential influence of CF collaterals on the interpretation of the results.
A direct activation of CN neurons by antidromic spikes in CF collaterals cannot be ruled out. However, we believe that this effect will not be substantial. The activation of the multi-synaptic pathway that we describe in this study is more likely to require a strong nudge as resulting from synchronized Purkinje cell input and subsequent rebound activation in CN neurons (PMID: 22198670), rather than small-amplitude input provided by CF collaterals (PMID: 27047344). A requirement for CF/PC synchronization would also set a threshold for activation of this suppressive pathway.
(3) The rationale behind the plasticity induction protocol for RWS+CF (50 ms light pulses at 1 Hz during 5 min of RWS, with a 45 ms delay relative to the onset of whisker stimulation) is unclear.
a) The authors state that 1 Hz was chosen to match the spontaneous CF firing rate (line 107); however, they also introduced a delay to mimic the CF response to whisker stimulation (line 108). This is confusing, and requires further clarification, specifically, whether the protocol was designed to reproduce spontaneous or sensory-evoked CF activity.
This protocol was designed to mimic sensory-evoked CF activity as reported in Bosman et al (J. Physiol. 588, 2010; PMID: 20724365).
b) Was the timing of delivering light pulses constant or random? Given the stochastic nature of CF firing, randomly timed light pulses with an average rate of 1Hz would be more physiologically relevant. At the very least, the authors should provide a clear explanation of how the stimulation timing was implemented.
Light pulses were delivered at a constant 1 Hz. Our goal was to isolate synchrony as the variable distinguishing sensory-evoked from spontaneous CF activity; additionally varying stochasticity, rate, or amplitude would have confounded this. Future studies could explore how these additional parameters shape S1 responses.
(4) CF activation modulates inhibitory interneurons in the S1 cortex (Figure 2): responses of interneurons in S1 to whisker stimulation were enhanced upon CF coactivation (Figure 2C), and these neurons were predominantly SST- and PV-positive interneurons (Figure 2H, I). In contrast, VIP-positive neurons were suppressed only in the late time window of 650-850 ms (Figure 2G). If the authors' hypothesis-that the activity of VIP neurons regulates SST- and PVneuron activity during RWS+CF-is correct, then the activity of SST- and PV-neurons should also be increased during this late time window. The authors should clarify whether such temporal dynamics were observed or could be inferred from their data.
Yes, we see a significant activity increase in PV neurons in this late time window (see updates to Data S2). Activity was also increased in SST neurons, though this did not reach statistical significance (Data S2). One reason might be that – given the small effect size overall – such an effect would only be seen in paired recordings. Chemogenetic activity modulation in VIP neurons, which provides a more crude test, shows, however, that SST- and PV-positive interneurons are indeed regulated via inhibition from VIP-positive interneurons (Fig. 5).
(5) Transsynaptic tracing from CN nicely identified zona incerta (ZI) neurons and their axon terminals in both POm and S1 (Figure 6 and Figure S7).
a) Which part of the CN (medial, interposed, or lateral) is involved in this pathway is unclear.
We used a dual-injection transsynaptic tracing approach to specifically label the outputs of ZI neurons that receive input from the deep cerebellar nuclei. The anterograde viral vector injected into the CN is unlabeled (no fluorophore) and therefore, it is not possible to reliably assess the extent of viral spread in those experiments as performed. However, we have previously performed similar injections into the deep cerebellar nuclei and post hoc histology suggest all three nuclei will have at least some viral expression (Koster and Sherman, 2024). Due to size and injection location, we will mostly have reached the lateral (dentate) nuclei, but cannot exclude partial transsynaptic tracing from the interposed and medial nuclei.
b) Were the electrophysiological properties of these ZI neurons consistent with those of PV neurons?
Although most recorded cells demonstrated electrophysiological properties consistent with PV+ interneurons in other brain regions (i.e. fast spiking, narrow spike width, non-adapting; see Tremblay et al., 2016), interneuron subtypes in the ZI have been incompletely characterized, with SST+ cells showing similar features to those typically associated with PV+ cells (if interested, compare Fig. 4 in DOI: 10.1126/sciadv.abf6709 vs. Fig. S10 in https://doi.org/10.1016/j.neuron.2020.04.027). Therefore, we did not attempt to delineate cell identity based on these characteristics.
c) There appears to be a considerable number of axons of these ZI neurons projecting to the S1 cortex (Figure S7C). Would it be possible to estimate the relative density of axons projecting to the POm versus those projecting to S1? In addition, the authors should discuss the potential functional role of this direct pathway from the ZI to the S1 cortex.
An absolute quantification is difficult to provide based on the images that we obtained. However, any crude estimate would indicate the relative density of projections to POm is higher than the density of projections to S1 (this is apparent from the images themselves). While the anatomical and functional connections from POm to S1 have been described in detail (Audette et al., 2018), this is not the case for the direct projections to ZI. A direct ZI to S1 projection would potentially involve a different recruitment of neurons in the S1 circuit. Any discussion on the specific consequences of the activation of this direct pathway would be purely speculative.
Reviewer #2 (Public review):
Summary:
The authors examined long-distance influence of climbing fiber (CF) signaling in the somatosensory cortex by manipulating whiskers through stimulation. Also, they examined CF signaling using two-photon imaging and mapped projections from the cerebellum to the somatosensory cortex using transsynaptic tracing. As a final manipulation, they used chemogenetics to perturb parvalbumin-positive neurons in the zona incerta and recorded from climbing fibers.
Strengths:
There are several strengths to this paper. The recordings were carefully performed, and AAVs used were selective and specific for the cell types and pathways being analyzed. In addition, the authors used multiple approaches that support climbing fiber pathways to distal regions of the brain. This work will impact the field and describes nice methods to target difficult-to-reach brain regions, such as the inferior olive.
Weaknesses:
There are some details in the methods that could be explained further. The discussion was very short and could connect the findings in a broader way.
In the revised manuscript, we provide more methodological details, as requested. We provided as simple as possible explanations in the discussion, so as not to bias further investigations into this novel phenomenon. In particular, we avoid an extended discussion of the gating effect of CF activity on S1 plasticity. While this is the effect on plasticity specifically observed here, we believe that the consequences of CF signaling on S1 activity may entirely depend on the contexts in which CF signals are naturally recruited, the ongoing activity of other brain regions, and behavioral state. Our key finding is that such modulation of neocortical plasticity can occur. How CF signaling controls plasticity of the neocortex in all contexts remains unknown, but needs to be thoughtfully tested in the future.
Reviewer #3 (Public review):
Summary:
The authors developed an interesting novel paradigm to probe the effects of cerebellar climbing fiber activation on short-term adaptation of somatosensory neocortical activity during repetitive whisker stimulation. Normally, RWS potentiated whisker responses in pyramidal cells and weakly suppressed them in interneurons, lasting for at least 1h. Crusii Optogenetic climbing fiber activation during RWS reduced or inverted these adaptive changes. This effect was generally mimicked or blocked with chemogenetic SST or VIP activation/suppression as predicted based on their "sign" in the circuit.
Strengths:
The central finding about CF modulation of S1 response adaptation is interesting, important, and convincing, and provides a jumping-off point for the field to start to think carefully about cerebellar modulation of neocortical plasticity.
Weaknesses:
The SST and VIP results appeared slightly weaker statistically, but I do not personally think this detracts from the importance of the initial finding (if there are multiple underlying mechanisms, modulating one may reproduce only a fraction of the effect size). I found the suggestion that zona incerta may be responsible for the cerebellar effects on S1 to be a more speculative result (it is not so easy with existing technology to effectively modulate this type of polysynaptic pathway), but this may be an interesting topic for the authors to follow up on in more detail in the future.
Our interpretation of the anatomical and physiological findings is that a pathway via the ZI is indeed critical for the observed effects. This pathway also represents perhaps the most direct pathway (i.e. least number of synapses connecting the cerebellar nuclei to S1). However, several other direct and indirect pathways are plausible as well and we expect distinct activation requirements and consequences for neurons in the S1 circuit. These are indeed interesting topics for future investigation.
Recommendations for the authors:
Reviewer #1 (Recommendations for the authors):
(1) Line 77: "CF transients" is not a standard or widely recognized term. Please use a more precise expression, such as "CF-induced calcium transients."
We now avoid the use of the term “CF transients” and replaced it with “CF-induced calcium transients.”
(2) Titer of AAVs injected should be provided.
AAV titers have been included in an additional data table (Data S9).
(3) Several citations to the figures are incorrect (for example, "Supplementary Data 2a (Line 398)" does not exist).
We apologize for the mistakes in this version of the article. Incorrect citations to the figures have been corrected.
(4) Line 627-628: "The tip of the patch cable was centered over Crus II in all optogenetic stimulation experiments." The stereotaxic coordinate of the tip position should be provided.
The stereotaxic coordinate of the tip position has been provided in the methods.
(5) Line 629: "Blue light pulses were delivered with a 470 nm Fiber-Coupled LED (Thorlabs catalog: M470F3)." The size of the light stim and estimated power density (W/mm^2) at the surface of the cortex should be provided.
The spot size and estimated power density at the surface of the cortex has been provided in the methods.
(6) Line 702-706: References for DCZ should be cited.
We now cited Nagai et al, Nat. Neurosci. 23 (2020) as the original reference.
(7) Two-photon image processing (Line 807-809): The rationale for normalizing ∆F/F traces to a pre-stimulus baseline is unclear because ∆F/F is, by definition, already normalized to baseline fluorescence: (Ft-F0)/F0. The authors should clarify why this additional normalization step was necessary and how it affected the interpretation of the data.
A single baseline fluorescence value (F₀) was computed for each neuron across the entire recording session, which lasted ~120-minutes. However, some S1 neurons exhibit fluctuations in baseline fluorescence over time—often related to locomotive activity or spontaneous network oscillations—which can obscure stimulus-evoked changes. To isolate fluorescence changes specifically attributable to whisker stimulation, we normalized each ∆F/F trace to the prestimulus baseline for that trial. This additional normalization allowed us to quantify potentiation or depression of sensory responses themselves, independently of spontaneous oscillations or locomotion-related changes in the ongoing neural activity.
Reviewer #2 (Recommendations for the authors):
(1) Did the climbing fiber stimulation for Figure 1 result in any changes to motor activity? Can you make any additional comments on other behaviors that were observed during these manipulations?
Acute CF stimulation did not cause any changes in locomotive or whisking activity. The CF stimulation also did not influence the overall level of locomotion or whisking during plasticity induction.
(2) Figure 3B and F- it is very difficult to see the SST+ neurons. Can this be enhanced?
We linearly adjusted the brightness and contrast for the bottom images in Figure 3B and F to improve visualization of SST+ neurons. Note the expression of both hM3D(Gq) and hM4D(Gi) in SST+ neurons is sparse, which was necessary to avoid off-target effects.
(3) Can you be more specific about the subregions of cerebellar nuclei and cell types that are targeted in the tracing studies? Discussions of the cerebellar nuclei subregions are missing and would be interesting, as others have shown discrete pathways between cerebellar nuclei subregions and long-distance projections.
See our response to comment 5a from Reviewer 1 (copied again here): we used a dual-injection transsynaptic tracing approach to specifically label the outputs of ZI neurons that receive input from the deep cerebellar nuclei. The anterograde viral vector injected into the CN is unlabeled (no fluorophone) and therefore, it is not possible to reliably assess the extent of viral spread in those experiments as performed. However, we have previously performed similar injections into the deep cerebellar nuclei and post hoc histology suggest all three nuclei will have at least some viral expression (Koster and Sherman, 2024). Due to size and injection location, we will mostly have reached the lateral (dentate) nuclei, but cannot exclude partial transsynaptic tracing from the interposed and medial nuclei.
It would indeed be interesting to further investigate the effect of CFs residing in different cerebellar lobules, which preferentially target different cerebellar nuclei, on targets of these nuclei.
(4) Did you see any connection to the ventral tegmental area? Can you comment on whether dopamine pathways are influenced by CF and in your manipulations?
We did not specifically look at these pathways and thus are not able to comment on this.
(5) These are intensive surgeries, do you think glia could have influenced any results?
This was not tested and seems unlikely, but we cannot exclude such possibility.
(6) It is unclear in the methods how long animals were recorded for in each experiment. Can you add more detail?
Additional detail was added to the methods. Recordings for all experimental configurations did not last more than 120 minutes in total. All data were analyzed across identical time windows for each experiment.
(7) In the methods it was mentioned that recording length can differ between animals. Can this influence the results, and if so, how was that controlled for?
There was a variance in recording length within experimental groups, but no systematic difference between groups.
(8) I do not see any mention of animal sex throughout this manuscript. If animals were mixed groups, were sex differences considered? Would it be expected that CF activity would be different in male and female mice?
As mentioned in the Methods (Animals), mice of either sex were used. No sex-dependent differences were observed.
(9) Transsynaptic tracing results of the zona incerta are very interesting. The zona incerta is highly understudied, but has been linked to feeding, locomotion, arousal, and novelty seeking. Do you think this pathway would explain some of the behavioral results found through other studies of cerebellar lobule perturbations? Some discussion of how this brain region would be important as a cerebellar connection in animal behavior would be interesting.
Since the multi-synaptic pathway from the cerebellum to S1 involves several brain regions with their own inputs and modulatory influences, it seems plausible to assume that behaviors controlled by these regions or affecting signaling pathways that regulate them would show some level of interaction. Our study does not address these interactions, but this will be an interesting question to be addressed in future work.
Reviewer #3 (Recommendations for the authors):
General comments on the data presentation:
I'm not a huge fan of taking areas under curves ('AUC' throughout the study) when the integral of the quantity has no physical meaning - 'normalizing' the AUC (1I,L etc) is even stranger, because of course if you instead normalize the AUC by the # of data points, you literally just get the mean (which is probably what should be used instead).
Indeed, AUC is equal to the average response in the time window used, multiplied by the window duration (thus, AUC is directly proportional to the mean). We choose to report AUC, a descriptive statistic, rather than the mean within this window. In 1I and L, we normalize the AUC across animals, essentially removing the variability across animals in the ‘Pre’ condition for visualization. Note the significance of these comparisons are consistent whether or not we normalize to the ‘Pre’ condition (non-normalized RWS data in I shows a significant increase in PN activity, p = 0.0068, signrank test; non-normalized RWS+CF data in I shows a significant decrease in PN activity, p = 0.0135, paired t-test; non-normalized RWS data in L shows a significant decrease in IN activity, p <0.001, paired t-test; non-normalized RWS+CF data in L shows no significant change in IN activity, p = 0.7789, paired t-test).
I think unadorned bar charts are generally excluded from most journals now. Consider replacing these with something that shows the raw datapoints if not too many, or the distribution across points.
We have replaced bar charts with box plots and violin plots. We have avoided plotting individual data points due to the quantity of points.
In various places, the statistics produce various questionable outcomes that will draw unwanted reader scrutiny. Many of the examples below involve tiny differences in means with overlapping error bars that are "significant" or a few cases of nonoverlapping error bars that are "not significant." I think replacing the bar charts may help to resolve things here if we can see the whole distribution or the raw data points. As importantly, I think a big problem is that the statistical tests all seem to be nonparametric (they are ambiguously described in Table S3 as "Wilcoxon," which should be clarified, since there is an unpaired Wilcoxon test [rank sum] and a paired Wilcoxon test [sign rank]), and thus based on differences in the *median* whereas the bar charts are based on the *mean* (and SEM rather than MAD or IQR or other medianappropriate measure of spread). This should be fixed (either change the test or change the plots), which will hopefully allay many of the items below.
We thank the reviewer for this important point. As mentioned in the Statistics and quantification section, Wilcoxon signed rank tests were used for non-normal data. We have replaced the bar charts with box plots which show the IQR and median, which indeed allays may of the items below.
Here are some specific points on the statistics presentation:
(1) 1G, the test says that following RWS+CF, the decrease in PN response is not significant. In 1I, the same data, but now over time, shows a highly significant decrease. This probably means that either the first test should be reconsidered (was this a paired comparison, which would "build in" the normalization subsequently used automatically?) or the second test should be reconsidered. It's especially strange because the n value in G, if based on cells, would seem to be ~50-times higher than that in I if based on mice.
In Figure 1G, the analysis tests whether individual pyramidal neurons significantly changed their responses before vs. after RWS+CF stimulation. This is a paired comparison at the single-cell level, and here indicates that the average per-neuron response did not reliably decrease after RWS+CF when comparing each cell’s pre- and post-values directly. In contrast, Figure 1I examines the same dataset analyzed across time bins using a two-way ANOVA, which tests for effects of time, group (RWS vs. RWS+CF), and their interaction. The analysis showed a significant group effect (p < 0.001), indicating that the overall level of activity across all time points differed between RWS and RWS+CF conditions. The difference in significance between these two analyses arises because the first test (Fig. 1G) assesses within-neuron changes (paired), whereas the second test (Fig. 1I) assesses overall population-level differences between groups over time (independent groups). Thus, the tests address related but distinct questions—one about per-cell response changes, the other about how activity differs across experimental conditions.
(2) 1J RWS+CF then shows a much smaller difference with overlapping error bars than the ns difference with nonoverlapping errors in 1G, but J gets three asterisks (same n-values).
Bar graphs have been replaced with box plots.
(3) 1K, it is very unclear what is under the asterisk could possibly be significant here, since the black and white dots overlap and trade places multiple times.
See response to point 1. A significant group effect will exist if the aggregate difference across all time bins exceeds within-group variability. The asterisk therefore reflects a statistically significant main group effect (RWS versus RWS+CF) rather than differences at any single time point. Note, however, the very small effect size here.
(4) 2B, 2G, 2H, 2I, 3G, 3H, 5C etc, again, significance with overlapping error bars, see suggestions above.
Bar graphs have been replaced with box plots.
(5) Time windows: e.g., L149-153 / 2B - this section reads weirdly. I think it would be less offputting to show a time-varying significance, if you want to make this point (there are various approaches to this floating around), or a decay rate, or something else.
Here, we wanted to understand the overall direction of influence of CFs on VIP activity. We find that CFs exert a suppressive effect on VIP activity, which is statistically significant in this later time window. The specific effect of CF modulation on the activity of S1 neurons across multiple time points will be described in more detail in future investigations.
(6) 4G, 6I, these asterisks again seem impossible (as currently presented).
Bar graphs have been replaced with box plots.
The writing is in generally ok shape, but needs tightening/clarifying:
(1) L45 "mechanistic capacity" not clear.
We have simplified this term to “capacity.” We use the term here to express that the central question we pose is whether CF signals are able to impact S1 circuits. We demonstrate CF signals indeed influence S1 circuits and further describe the mechanism through which this occurs, but we do not yet know all of the natural conditions in which this may occur. We feel that “capacity” describes the question we pose -- and our findings -- very well.
(2) L48-58 there's a lot of material here, not clear how much is essential to the present study.
We would like to give an overview of the literature on instructive CF signaling within the cerebellum. Here, we feel it is important to describe how CFs supervise learning in the cerebellum via coincident activation of parallel fiber inputs and CF inputs. Our results demonstrate CFs have the capacity to supervise learning in the neocortex in a similar manner, as coincident CF activation with sensory input modulates plasticity of S1 neurons.
(3) L59 "has the capacity to" maybe just "can".
This has been adopted. We agree that “can” is a more straightforward way of saying “has the capacity to” here. In this sentence, “can” and “has the capacity to” both mean a general ability to do something, without explicit knowledge about the conditions of use.
(4) L61-62 some of this is circular "observation that CF regulates plasticity in S1..has consequences for plasticity in S1".
We now changed this to read “…consequences for input processing in S1.”
(5) L91 "already existing whisker input" although I get it, strictly speaking, not clear what this means.
This sentence has been reworded for clarity.
(6) L94 "this form of plasticity" what form?
Edited to read “sensory-evoked plasticity.”
(7) L119 should say "to test the".
This has been corrected.
(8) L120 should say "well-suited to measure receptive fields".
We agree; this wording has been adopted.
(9) L130 should say "optical imaging demonstrated that receptive field".
This has been adopted.
(10) L138, the disclaimer is helpful, but wouldn't it be less confusing to just pick a different set of terms? Response potentiation etc.
Perhaps, but we want to stress that components of LTP and LTD (traditionally tested using electrophysiological methods to specifically measure synaptic gain changes) can be optically measured as long as it is specified what is recorded.
(11) L140, this whole section is not very clear. What was the experiment? What was done and how?
The text in this section has been updated.
(12) L154, 156, 158, 160, 960, what is a "basic response"? Is this supposed to contrast with RWS? If so, I would just say "we measured the response to whisker stimulation without first performing RWS, and compared this to the whisker stimulation with simultaneous CF activation."
What we meant by “basic response” was the acute response of S1 neurons to a single 100 ms air puff. Here, we indeed measured the acute responses of S1 neurons to whisker stimulation (100 ms air puff) and compared them to whisker stimulation with simultaneous CF activation (100 ms air puff with a 50 ms light pulse; the light pulse was delayed 45 ms with respect to the air puff). This paragraph has been reworded for clarity.
(13) L156 "comprised of a majority" unclear. You mean most of the nonspecific IN group is either PV or SST?
Yes, that was meant here. This paragraph has been reworded for clarity.
(14) L165 tense. "are activated" "we tested" prob should be "were activated."
This sentence was reworded.
(15) L173 Not requesting additional experiments, but demonstrating that the effect is mimicked by directly activating SST or suppressing VIP questions the specificity of CF activation per se, versus presumably many other pathways upstream of the same mechanisms, which might be worth acknowledging in the text.
We indeed observe that directly activating SST or suppressing VIP neurons in S1 is sufficient to mediate the effect of CF activation on S1 pyramidal neurons, implicating SST and VIP neurons as the local effectors of CF signaling. In the text, we wrote “...the notion of sufficiency does not exclude potential effects of plasticity processes elsewhere that might well modulate effector activation in this context and others not yet tested.” Here, we mean that CFs are certainly not the only modulators of the inhibitory network in S1. One example we highlight in the discussion is that projections from M1 are known to modulate this disinhibitory VIP-to-SST-to-PN microcircuit in S1. We conclude from our chemogenetic manipulation experiments that CFs ultimately have the capacity to modulate S1 interneurons, which must occur indirectly (either through the thalamus or “upstream” regions as this reviewer points out). The fact that many other brain regions may also modulate the interneuron network in S1 -- or be modulated by CF activity themselves -- only expands the capacity of CFs to exert a variety of effects on S1 neurons in different contexts.
(16) L247 "induced ChR2" awkward.
We changed this to read “we expressed ChR2.”
(17) 6C, what are the three colors supposed to represent?
We apologize for the missing labels in this version of the manuscript. Figure 6C and the figure legend have been updated.
-
-
social-media-ethics-automation.github.io social-media-ethics-automation.github.io
-
21.2. Ethics in Tech# In the first chapter of our book we quoted actor Kumail Nanjiani on tech innovators’ lack of consideration of ethical implications of their work. Of course, concerns about the implications of technological advancement are nothing new. In Plato’s Phaedrus [u1] (~370BCE), Socrates tells (or makes up[1]) a story from Egypt critical of the invention of writing: Now in those days the god Thamus was the king of the whole country of Egypt, […] [then] came Theuth and showed his inventions, desiring that the other Egyptians might be allowed to have the benefit of them; […] [W]hen they came to letters, This, said Theuth, will make the Egyptians wiser and give them better memories; it is a specific both for the memory and for the wit. Thamus replied: […] this discovery of yours will create forgetfulness in the learners’ souls, because they will not use their memories; they will trust to the external written characters and not remember of themselves. The specific which you have discovered is an aid not to memory, but to reminiscence, and you give your disciples not truth, but only the semblance of truth; they will be hearers of many things and will have learned nothing; they will appear to be omniscient and will generally know nothing; they will be tiresome company, having the show of wisdom without the reality. In England in the early 1800s, Luddites [u2] were upset that textile factories were using machines to replace them, leaving them unemployed, so they sabotaged the machines. The English government sent soldiers to stop them, killing and executing many. (See also Sci-Fi author Ted Chiang on Luddites and AI [u3]) Fig. 21.1 The start of an xkcd comic [u4] compiling a hundred years of complaints about how technology has speed up the pace of life. (full transcript of comic available at explainxkcd [u5])# Inventors ignoring the ethical consequences of their creations is nothing new as well, and gets critiqued regularly: Fig. 21.2 A major theme of the movie Jurassic Park (1993) [u6] is scientists not thinking through the implications of their creations.# Fig. 21.3 Tweet parodying how tech innovator often do blatantly unethical things [u7]# Many people like to believe (or at least convince others) that they are doing something to make the world a better place, as in this parody clip from the Silicon Valley show [u8] (the one Kumail Nanjiani was on, though not in this clip): But even people who thought they were doing something good regretted the consequences of their creations, such as Eli Whitney [u9] who hoped his invention of the cotton gin would reduce slavery in the United States, but only made it worse, or Alfred Nobel [u10] who invented dynamite (which could be used in construction or in war) and decided to create the Nobel prizes, or Albert Einstein regretting his role in convincing the US government to invent nuclear weapons [u11], or Aza Raskin regretting his invention infinite scroll. [1] In response to Socrates’ story, his debate partner Phaedrus says, “Yes, Socrates, you can easily invent tales of Egypt, or of any other country.”
What stood out to me is how every generation thinks new technology is going to ruin everything, whether it’s writing, machines, or even social media today. It makes me wonder if our fears about tech—like AI or algorithms—are actually about the tech itself or more about us not wanting to adapt. At the same time, I don’t think these concerns are pointless. The examples of Eli Whitney and Alfred Nobel show that even good intentions can lead to huge negative consequences. So I feel like the real issue isn’t whether technology is good or bad, but whether the people creating it are actually thinking ahead about the impact. Honestly, most of the time it feels like they’re not, especially when companies rush to build something new just because they can. It makes me question: what current tech are we going to look back on in 20 years and say, “Wow, that messed things up more than we expected”?
-
-
www.biorxiv.org www.biorxiv.org
-
Note: This response was posted by the corresponding author to Review Commons. The content has not been altered except for formatting.
Learn more at Review Commons
Reply to the reviewers
Manuscript number: RC-2025-03174
Corresponding author(s): Cristina, Tocchini and Susan, Mango
1. General Statements
We thank the reviewers for their thoughtful and constructive comments. We were pleased that the reviewers found our study “rigorous”, “well presented”, “technically strong”, and “novel”. We are also grateful for their recognition that our work identifies a function for a HOT region in gene regulation and provides new insights into the role of the uHOT in controlling dlg-1 expression.
Point-by-point description of the revisions
We have addressed the reviewers’ concerns by clarifying and refining the text, particularly regarding the intron 1 results, improving the quantitation and statistical analyses, and making adjustments and additions to text and figures.
Specific responses to each point are provided below in blue.
Reviewer #1
-
- The results fully support the authors conclusions regarding the significant role of the upstream HOT region ("uHOT") with strong fluorescence activity and substantial phenotypic effects (i.e., the animals have very low brood sizes and rarely progress through hatching). This data is well presented and technically well done.* Thank you.
-
In my view, their conclusions regarding the intronic HOT region are speculative and unconvincing. See below for main criticisms.*
We agree, and have made changes throughout the manuscript to make this point clearer. Specifically, we contextualize the role of intron 1 as a putative enhancer in reporter assays, but not in endogenous, physiological conditions. Some examples are:
Abstract: “(…) In contrast, the intronic region displays weak enhancer-like activity when tested in transcriptional reporter assays but is dispensable in transcriptional control when studied at the endogenous locus. Our findings reveal how HOT regions contribute to gene regulation during animal development and illustrate how regulatory potential identified in isolated contexts can be selectively deployed or buffered within the native genomic architecture.”
Background: “(…) The HOT region in the first intron possesses weak transcriptional capabilities that are restricted to epidermal cells as observed in transcriptional reporters, but seem to not be employed in physiological contexts.” As it will become clear reading this updated version of the manuscript, we cannot exclude at present a functional role during non-physiological conditions (e.g., stress)
Results and discussion: “(…) This is in contrast with what the reporter experiments showed, where intron 1 alone was permissive for transcription and slightly enhanced the FL transgene expression levels (Figure 1F,G and S4). (…)”
Other changes can be found highlighted in yellow in the manuscript.
- Furthermore, their conclusions about interactions between the two tested regions is speculative and they show no strong evidence for this claim.*
We thank the reviewer for raising this concern. To avoid overstating our conclusions, we now frame the potential interaction between the two studied HOT regions strictly in the context of previously published ARC-C data (Huang et al., 2022). We clarify in the revised text that these interactions have been observed in earlier work during larval stages (Huang et al., 2022), but remain to be validated during embryogenesis, and we present them solely as contextual information rather than as a central conclusion.
In Results and discussion section we wrote: “(…) Although the presence of a fountain at this locus remains to be confirmed during embryogenesis, Accessible Region Conformation Capture (ARC-C), a method that maps chromatin contacts anchored at accessible regulatory elements, showed that the putative HOT region interacts with other DNA sequences, including the first intron of dlg-1 (1). (…)”
* The authors claim that not all the phenotypic effects seen from deleting the uHOT region are specific to the dlg-1 gene. This is an interesting model, but the authors show essentially no data to support this or any explanation of what other gene might be regulated.*
We appreciate the reviewer’s comment and have revised the manuscript to ensure that the possibility of additional regulatory effects from the uHOT region is presented as a hypothesis rather than a claim. Our study was designed to investigate HOT-region–based transcriptional regulation rather than chromatin interactions, and we now make this scope more explicit in the text. The revised discussion highlights that, although ARC-C data suggest the uHOT region may contact other loci, the idea that these interactions contribute to the observed phenotypes remains speculative and will require dedicated future work.
In Results and discussion section we wrote: “(…) Because, as previously shown, the upstream HOT region exhibits chromatin interactions with other genomic loci (1), its depletion might affect gene expression of beyond dlg-1 alone. An intriguing hypothesis is that these phenotypes do not arise only from the reduction in dlg-1 mRNA and DLG-1 protein levels, but also from synergistic, partial loss-of-function phenotypes involving other genes (24). (…)”
* Finally, some of the hypotheses in the text could be more accurately framed by the authors. They claim HOT regions are often considered non-functional (lines 189-191). Also, they claim that correct expression levels and patterning is usually regulation by elements within a few hundred basepairs of the CDS (lines 78-80). These claims are not generally accepted in the field, despite a relatively compact genome. Notably, both claims were tested and disproven by Chen et al (2014), Genome Research, where the authors specifically showed strong transcriptional activity from 10 out of 10 HOT regions located up to 4.7 kb upstream of their nearest gene. Chen et al. 2014 is cited by Tocchini et al. and it is, therefore, surprisingly inconsistent with the claims in this manuscript.*
We thank the reviewer for this comment and have revised the text to clarify our intended meaning and avoid framing discussion points as absolute claims. We changed “often” to “frequently” in both sentences so that they better reflect general trends rather than universal rules.
The revised text now reads: “Controversially, C. elegans sequences that dictate correct expression levels and patterning are frequently located within a few hundred base-pairs (bp) (maximum around 1,000–1,500 bp) from a gene’s CDS (3,13–15),”;
And: “HOT regions in C. elegans, as well as other systems, have been predominantly associated with promoters and were frequently considered non-functional or simply reflective of accessible chromatin (25).”
Regarding the comparison to Chen et al., 2014, we note that their reporters did not include a reference baseline for “strong” transcriptional activity, and only five of the ten tested HOT regions were located more than 1.5 kb from the nearest TSS. Therefore, our phrasing is consistent with their findings while describing general trends observed in the C. elegans genome rather than absolute rules. We have also ensured that these sentences are presented as discussion points rather than definitive claims. We hope these revisions make the framing and context clearer to the reader. The fluorescence expression from the intronic HOT region is not visible by eye and the quantification shows very little expression, suggestive of background fluorescence. Although the authors show statistical significance in Figure 1G, I would argue this is possibly based on inappropriate comparisons and/or a wrong choice statistical test. The fluorescence levels should be compared to a non-transgenic animal and/or to a transgenic animal with the tested region shuffled but in an equivalent
We understand the reviewer’s concern regarding the low fluorescence levels observed for the intronic HOT reporter. To address this, we have now included a Figure S4 with higher-exposure versions of the embryos shown in Figure 1. These panels confirm that the nuclear signal is genuine: embryos without a functional transcriptional transgene do not display any comparable fluorescence, aside from the characteristic cytoplasmic granules associated with embryonic autofluorescence. Similar reference images have also been added to Figure S3 to clarify the appearance of autofluorescence under the same imaging conditions.
Regarding the quantitation analyses, as suggested by the reviewers, we now consistently quantify fluorescence by calculating the mean intensity for each embryo (biological replicates) and performing statistical analyses on these values. This approach ensures that the statistical tests are applied to independent biological measurements.
* I would suggest the authors remove their claims about the intronic enhancer and the interaction between the two regions. And I would suggest softening the claims about the uHOT regulation of another putatitive gene.*
We have revised the manuscript to avoid definitive claims regarding the presence of an interaction between the two studied HOT regions. These points are now presented strictly as hypotheses within the discussion, suggested by previously published ARC-C data rather than by our own experimental evidence. Likewise, we have softened our statements regarding the possibility that the uHOT region may regulate additional gene(s). This idea is now framed as a speculative model that will require dedicated future studies, rather than as a conclusion of the present work. Quotes can be found in the previous points (#3 and #4) raised by Reviewer 1.
* The authors would need to demonstrate several things to support their current claims. The major experiments necessary are:*
-
- Insert single-copy transgene with a minimal promoter and the intronic sequence scrambled to generate a proper baseline control. It is very possible that the intronic sequence does drive some expression, but the current control is not appropriate for statistical comparison (e.g., only the transgene with intron 1 contains the minimal promoter from pes-10, which may have baseline transcriptional activity even without the intron placed in front of the transgene).* We thank the reviewer for this suggestion. We agree that a scrambled-sequence control can be informative in some contexts; however, in this case we believe the existing data already address the concern. In our dataset, all uHOT reporter constructs—each containing the same minimal promoter—show consistent background levels in the absence of regulatory input, providing an internal baseline for comparison. For this reason, we consider the current controls sufficient to interpret the effects of the intronic region in reporter assays.
In general, the minimal Δpes-10 promoter is specifically designed to have negligible basal transcriptional activity on its own, and this property has been extensively validated in previous studies (reference included in the revised manuscript).
* It is not very clear why the authors did not test intron 1 within the H2B of the transgene and just the minimal promoter in front of the transgene, but only in the context of the full-length promoter. The authors show a minor difference in expression levels for the full-length (FL) and full-length with intron 1 (FL-INT1) but show a large statistical differnce. The authors use an inappropriate statistical test (T-test) for this experiment and treat many datapoints from the same embryo as independent, which is clearly not the case. Even minor differences in staging, transgene silencing in early development, or variability would potentially bias their data collection.*
We thank the reviewer for this comment. Our goal was to assess the potential contribution of intron 1 in two complementary contexts: (i) on its own, upstream of a minimal promoter, to test whether it can in principle support transcription, and (ii) within the full-length promoter construct, which more closely reflects the endogenous configuration. For this reason, we did not generate an additional construct placing intron 1 within the H2B reporter driven only by the minimal promoter, as we considered this redundant with the information provided by the existing INT1 and FL-INT1 reporters.
Regarding the statistical analysis, we agree that treating multiple measurements from the same embryo as independent is not appropriate. In the revised manuscript, we now use the mean fluorescence intensity per embryo as a single biological replicate and perform all statistical tests on these independent values. This approach avoids pseudo-replication and ensures that the analysis is robust to variability in staging or transgene behavior. The conclusions remain the same.
* The authors claim, based on ARC-C data previously published by their lab (Huang et al. 2022) that the dlg-1 HOT region interacts with "other" genomic regions. This is potentially interesting but the evidence for this should be included in the manuscript itself, perhaps by re-analyzing data from the 2022 manuscript?*
We thank the reviewer for this suggestion. The chromatin-interaction data referred to in the manuscript originate from the work of Huang et al., 2022, published by the Ahringer lab. As these ARC-C datasets are already publicly available and thoroughly analyzed in the original publication, we felt that reproducing them in our manuscript was not necessary for supporting the limited contextual point we make. Our intent is simply to note that previous work reported contacts between the uHOT region and additional loci. To address the reviewer’s concern, we have revised the manuscript to make clear that we are referencing previously published ARC-C observations and that we do not present these interactions as new findings from our study.
For example, in Results and discussion section we wrote: “(…) Because, as previously shown, the upstream HOT region exhibits chromatin interactions with other genomic loci (1), its depletion might affect gene expression beyond dlg-1 alone. An intriguing hypothesis is that these phenotypes do not arise only from the reduction in dlg-1 mRNA and DLG-1 protein levels, but also from a synergistic, partial loss-of-function phenotypes involving other genes (24). (…)”
* The fluorescence quantification is difficult to interpret from the attached data file (Table S1). For the invidividual values, it is unclear how many indpendent experiments (different embryos) were conducted. The authors should clarify if every data value is from an independent embryo or if they used several values from the same embryo. If they did use several values from the same embryo, how did they do this? Did they take very cell? Or did they focus on specific cells? How did they ensure embryo staging?*
We thank the reviewer for pointing this out. To clarify the quantification procedure, we have expanded the description in the Methods section (“Live imaging: microscopy, quantitation, and analysis”). The revised text now specifies that each data point represents the normalized fluorescence value obtained from three nuclei (or five junctions, depending on the construct), all taken from the same anatomical positions across embryos. Two independent biological replicates were performed for each experiment, with each embryo contributing a single averaged value.
As noted in the figure legends, the specific nuclei used for quantification are indicated in each panel (with dashed outlines), and a reference nucleus marked with an asterisk allows unambiguous identification of the same positions across all conditions. We are happy to further refine this description if additional clarification is needed.
* The authors also do not describe how they validated single-copy insertions (partial transgene deletions in integrants are not infrequent and they only appear to use a single insertion for each strain). This should be described and or added as a caveat if no validation was performed.*
The authors also do not describe any validation for the CRISPR alleles, either deletions or insertion of the synthetic intron into dlg-1. How were accurate gene edits verified.
We thank the reviewer for highlighting the importance of validating the genetic constructs. We have now clarified this more explicitly in the revised Methods section and in Table S1. All single-copy transgene insertions and all CRISPR-generated alleles were verified by genotyping and Sanger sequencing to confirm correct integration and the absence of unintended rearrangements.
- *
I am not convinced the statistical analysis of the fluorescence data is correct. Unless the authors show that every datapoint in the fluorescence quantification is independent, then I would argue they vastly overestimate the statistical significance. Even small differences are shown to have "***" levels of significance, which does not appear empirically plausible.
We thank the reviewer for highlighting this point. To ensure that each data point represents an independent measurement, we now calculate the mean fluorescence per embryo (from three nuclei or five junctions) and use these per-embryo means as biological replicates for statistical testing. Two independent experiments were performed for each condition. Statistical differences were evaluated using a one-tailed t-test on the per-embryo means, as indicated in the revised Methods section.
After this adjustment, the differences remain statistically significant, although less extreme than in the initial analysis (now p * *
This study is so closely related to the Chen et al study, that I believe this study should be discussed in more detail to put the data into context.
We thank the reviewer for this suggestion. While we refer to Chen et al., 2014 as a relevant prior study for context, we believe that our work addresses distinct questions and experimental approaches. Specifically, our study focuses on HOT region-based transcriptional regulation in the dlg-1 locus and its functional dissection in vivo, which is conceptually and methodologically different from the scope of Chen et al., 2014 where the author tested the functionality of HOT region-containing promoters in the context of single-copy integrated transcriptional reporters. We hope this is clearer to the reader in the revised manuscript.
* Add H2B to the mNG in Figure 1 in order to understand where the first intron was inserted.*
We thank the reviewer for this suggestion. A schematic representation of the transgene is already provided above the corresponding images to indicate the location of the first intron.
For additional clarity, we have now added the following sentence in the main text: “In the other, intron 1 was inserted in the FL transgene within the H2B coding sequence (at position 25 from the ATG), preserving the canonical splice junctions with AG at the end of the first exon and a G at the beginning of the second exon, so that it acted as a bona fide intron (FL-INT1) (Figure 1F).”
This should help readers understand the placement of the intron without requiring modifications to the figure itself.__ __
Reviewer #2
1) The authors suggest that the region upstream of the dlg-1 gene is a HOT region. Although they highlight that other broad studies pick up this region as a HOT region, it would be good that the authors dive into the HOT identity of the region and characterize it, as it is a major part of their study. In addition to multiple TFs binding to the site, there are different criteria by which a region would be considered a HOT region. E.g. is there increased signal on this region in the IgG ChIP-seq tracks? Is the area CpG dense?
We thank the reviewer for this suggestion. In the manuscript and Figure S1, we show several features of HOT regions, including transcription factor binding and chromatin marks. To further characterize the dlg-1 uHOT region, we have added the following sentence to the text: “The conserved region is positioned approximately four Kb from the CDS of dlg-1 in a CpG-dense sequence (2), and is overlapping and bordered by chromatin marks typically found in enhancers (5,16).”
This addition provides additional evidence supporting the identity of the region as a HOT region, complementing the features already presented.
* 2) When describing the HOT region, they refer to Pol II binding as 'confirming its role as a promoter': non-promoter regions can also have Pol II binding, especially enhancers. Having binding of Pol II does not confirm its role as promoter. On the contrary, seeing the K27ac and K4me1 would point towards it being an enhancer.*
The sentence has been revised to clarify the interpretation of Pol II binding: “This HOT site also contains RNA Pol II peaks during embryogenesis (Figure S1C), supporting its role as a promoter or enhancer (9).” This wording avoids overinterpreting Pol II binding alone, while acknowledging that the HOT region may have both promoter and enhancer characteristics.
We would like to note that the relevant chromatin marks (H3K27ac and H3K4me1), which are indicative of enhancer activity, are described in the text: “(…) Specifically, it is enriched in acetylated lysine 27 (H3K27ac) and mono- and di-methylated lysine 4 of histone H3 (H3K4me1/2), and depleted from tri-methylated lysine 4 of histone H3 (H3K4me3) (Figure S1D) (5,16). (…)”
These changes clarify that the HOT region may have enhancer characteristics and avoid overinterpreting the Pol II signal.
* 3) In S1B, the authors show TF binding tracks. They also have a diagram of the region subsets (HOT1-4) that were later tested. What is their criteria for dividing the HOT region into those fragments? From looking at Fig S1, the 'proper' HOT region (ie. Where protein binding occurs) seems to be divided into two (one chunk as part of HOT3 and one chunk as part of HOT4). Can the authors comment on the effects of this division?*
To clarify the criteria for dividing the HOT region into subregions, we have added the following sentence to the main text: “The subregions were chosen taking into account (i) enrichment of putative TF binding sites (uHOT1 for PHA-4, uHOT2 for YAP-1 and NHR-25, uHOT3 for ELT-3, and uHOT4 for PHA-4 and others (e.g., ELT-1 and ELT-3)), (ii) Pol II binding peaks, and (iii) histone modification peaks (Fig. S1C,D).”
This description explains the rationale behind the division and clarifies why the HOT region was split into these four fragments for functional testing.
* 4) For the reporter experiments, the first experiments carry the histone H2B sequence and the second set of experiments (where the HOT region is dissected) carry a minimal promoter Δ*pes-10 (MINp). The results could be affected by the addition of these sequences. Is there a reason for this difference? Can the authors please justify it?
The difference in reporter design reflects the distinct goals of the two sets of experiments. The H2B sequence, coupled to mNG, is used as a coding sequence throughout the first part of the study (reporter analysis). This is commonly used to (i) concentrate the fluorescence signal (mNG) into nuclei (H2B) and (ii) be able to identify specific cells more accurately for quantitation reasons (intensity and consistency). The Δpes-10 promoter is instead used to analyze whether specific sequences possess enhancer potential: this promoter alone possesses the sequences that can allow transcription only in the presence of transcription factors that bind to the studied sequence placed upstream it.
To clarify this distinction in the manuscript, we have added the following sentence: “(…) Each region was paired with the minimal promoter Δpes-10 (MINp) (Figure 1D) and generated four transcriptional reporters. Δpes-10 is commonly used to generate transcriptional reporter aimed at assessing candidate regulatory enhancer sequences (20). The minimal promoter drives expression only when transcription factors bind to the tested upstream sequence and test enhancer activity. (…)”
5) Regarding the H2B sequence: ' 137: first intron [...] inserted in the FL transgene within the H2B sequence, acting as an actual intron (FL-INT1)': how was the location of the insertion chosen? Does it disrupt H2B? can it be that the H2B sequence contributed to dampening down the expression of mNG and disrupting it makes it stronger? It would be important to run the first experiments with minimal promoters and not with the H2B sequence.
The location of the intron insertion within the H2B coding sequence was chosen to preserve proper splicing and avoid disrupting H2B protein. We added the following sentence to clarify this point: “(…) In the other, the intron was inserted in the FL transgene within the H2B coding sequence (at position 25 from the ATG), preserving the canonical splice junctions with AG at the end of the first exon and a G at the beginning of the second exon, so that it acted as a bona fide intron (FL-INT1) (Figure 1F). (…)”
* 6) Have the authors explored the features of the sequences underlying the different HOT subregions? (e.g. running a motif enrichment analysis)? Is there anything special about HOT3 that could make it a functional region? It would be good to compare uHOT3 vs the others that do not drive the correct pattern. Since it's a HOT region, it may not have a special feature, but it is important to look into it.*
We thank the reviewer for this suggestion. To clarify the rationale for dividing the HOT region into four subregions, we have added the following sentence to the main text: “(…) The subregions were chosen taking into account (i) enrichment of putative TF binding sites (uHOT1 for PHA-4, uHOT2 for YAP-1 and NHR-25, uHOT3 for ELT-3, and uHOT4 for PHA-4 and others (e.g., ELT-1 and ELT-3)), (ii) Pol II binding peaks, and (iii) histone modification peaks (Fig. S1C,D). (…)”
While uHOT3 does not appear to possess unique sequence features beyond these general HOT-region characteristics, this approach allowed us to systematically test which fragments contribute to transcriptional activity and patterning.
7) For comparisons, the authors run t-tests. Is the data parametric? Otherwise, it would be more suitable to use a non-parametric test.
To ensure that each data point represents an independent biological replicate, we now calculate the mean fluorescence intensity per embryo and perform statistical tests on these per-embryo means. The data meet the assumptions of parametric tests, and we use a one-tailed t-test as indicated in the Methods.
* 1) The authors work with C. elegans embryos at comma stage, according to the methods section. It would be good if the authors mentioned it in the main text so that the reader is informed.*
Thanks for this suggestion. We added this sentence in the main text: “(…) Live imaging and quantitation analyses on embryos at the comma stage (used throughout the study for consistency purposes) showed (…)”.
* 2) 'Notably, the upstream HOT region is located more than four kilo-bases (Kb) away the CDS, and the one in the first intron contains enhancer sites, too.': what do they mean by 'enhance sites, too'. Is the region known as a functional enhancer? If so, could you please provide the reference?*
Here the clarification from the revised text: “(…) Notably, the upstream HOT region is located more than four kilo-bases (Kb) away the CDS, and the one in the first intron does not only contain two TSS but also three enhancer sites (8). (…)”
* 3) 'We hypothesized the upstream HOT region is the main driver of dlg-1 transcriptional regulation.': this sentence needs more reasoning. What led to this hypothesis? Is it the fact of seeing multiple TFs binding there? The chromatin marks?*
The reasoning behind the hypothesis is described in the preceding paragraph, and to make this connection clearer, we have revised the sentence to begin with: “Considering all of this information, we hypothesized the upstream HOT region is the main driver of dlg-1 transcriptional regulation. (…)”.
This change explicitly links the hypothesis to the observed TF binding and chromatin marks described above.
* 4) The labels of S1B are too wide, as if they have stretched the image. Could the authors please correct this?*
Yes, we agree with Reviewer 2. We corrected this.
* 5) This sentence does not flow with the rest of the text '84 - cohesins have been shown to organize the DNA in a way that active enhancers make contacts in the 3D space forming "fountains" detectable in Hi-C data (17,18).': is there a reason to explain this? I would remove it if not, as it can confuse the reader.*
We thank the reviewer for this comment. We agree that the sentence could potentially interrupt the flow; however, it is important for introducing the concept of “fountains” in 3D genome organization, which is necessary to understand the subsequent statement: “(…) Although the presence of a fountain at this locus remains to be confirmed during embryogenesis, Accessible Region Conformation Capture (ARC-C), a method that maps chromatin contacts anchored at accessible regulatory elements, showed that the putative HOT region interacts with other DNA sequences, including the first intron of dlg-1 (1). (…)”.
Therefore, we have retained this sentence to provide the necessary background for readers.
* 6) The authors mentioned that 'ARC-C data showed the putative HOT region interacts with other DNA sequences, including the first intron of dlg': have the authors analysed the data from the previous paper? A figure with the relevant data could illustrate this interaction so that the reader knows which specific region has been shown to interact with which. This would also bring clarity as to why they chose intron1 for additional experiments.*
We thank the reviewer for this suggestion. We have examined the relevant ARC-C data from the previous publication (Huang et al., 2022). However, as these results are already published, we do not feel it is necessary to reproduce them in our manuscript. The mentioning of these interactions is intended only to introduce the concept for discussion and to provide context for why intron 1 was considered in subsequent experiments
* 7) 'two deletion sequences spanning from the beginning (uHOT) or the end (Short) of the HOT region until the dlg-1 CDS': From the diagrams of the figure, I understand that uHOT has the distal region deleted, and the short HOT has the distal and the upstream regions deleted. Is this correct? Could you clarify this in the text? E.g. 'we designed two reporters - one containing the sequence starting at the HOT region and ending at the dlg-1 CDS, and the other without the HOT region, but rather starting downstream of it until the dlg-1 CDS'.*
To clarify the design of the reporters, we have revised the text as follows: “(…) To test this idea, we generated three single-copy, integrated transcriptional reporters carrying a histone H2B sequence fused to an mNeon-Green (mNG) fluorescent protein sequence under the transcriptional control of the following dlg-1 upstream regions: (i) a full-length sequence (“FL” = Distal + uHOT + Proximal sequences), (ii) one spanning from the beginning of the HOT region to the dlg-1 CDS (“uHOT” = uHOT + Proximal sequences), and (iii) one starting at the end of the HOT region and ending at the dlg-1 CDS (“Short” = Proximal sequence) (Figure 1A-C). (…)”
This description clarifies which parts of the upstream region are included in each reporter and matches the schematics in Figure 1.
* 8) 'Specifically, it spanned from bp 5,475,070 to 5,475,709 on chromosome X and removed HOT2 and HOT2 sequences' - this is unclear to me. What sequences are removed? HOT2 and 3?*
Thanks for spotting this typo. It has now been corrected.
* 9) 'ARC-C' is not introduced. Please spell out what this is. Accessible Region Conformation Capture (ARC-C). It would be helpful to include a sentence of what it is, as it will not be known by many readers.*
You are right, we changed into: “(…) Although the presence of a fountain at this locus remains to be confirmed during embryogenesis, Accessible Region Conformation Capture (ARC-C), a method that maps chromatin contacts anchored at accessible regulatory elements, showed that the putative HOT region interacts with other DNA sequences, including the first intron of dlg-1 (1). (...)”
* 10) Fig 1 B, diagram on the right: the H2B sequence is missing. I see that is indicated in the legend as part of mNG but this can be misleading. Could the authors add it to the diagram for clarification?*
Yes, you are right. We added this in the figure.__ __
Reviewer #3
The authors' claims are generally supported by the data, thoug the last sentence of the abstract was a bit overstated. They state that they "reveal the function of HOT regions in animals development...."; it would be more accurate to state that they linked the role of an upstream HOT region to dlg-1 regulation, and their findings hint that this element could have additional regulatory functions. The authors can either temper their conclusions or try RNA-seq experiments to find additional genes that are misregulated by the delta-uHOT deletion allele. [OPTIONAL]. Another [OPTIONAL] experiment that would strengthen the claims is to perform RNAi knockdown or DLG-1 protein depletion and link that to phenotype to show that the dlg-1 mRNA and DLG-1 protein changes seen in the uHOT mutant do not explain the lethality observed.
We thank the reviewer for this comment. We have studied HOT region function in the context of a model organism, C. elegans; therefore, we believe that describing our findings as revealing a function of HOT regions in animal development is accurate. The sentence aims at noting that these observations may provide broader insights into HOT region regulation. We changed the last sentence of the abstract into: “(…) Our findings reveal how HOT regions contribute to gene regulation during animal development and illustrate how regulatory potential identified in isolated contexts can be selectively deployed or buffered within the native genomic architecture. (…)”.
We note that RNA-seq is beyond the scope of this study; our discussion of potential effects on other genes is intended only as a hypothesis for future work. RNAi of dlg-1 has been previously reported and is cited in the manuscript, providing context for the phenotypes observed and discussed.
-
* When printed out I cannot read what the tracks are in Fig S1. Adding larger text to indicate what those tracks are is necessary.* Yes, you are right. We changed this in the figure.
-
*
-
Line 79. I would change the word "usually" to "frequently" in the discussion about regulatory element position. While promoters ranging from a few hundred to 2000 basepairs are frequently used, there are numerous examples where important enhancers can be further away.*
Corrected.
* Line 93-95. The description of the reporters was very confusing. When referring to the deletion sequences it sounds like that is what is missing rather than what is included. Rather, if I understand correctly the uHOT is the sequence from the start of the uHOT to the CDS and Short starts at the end of uHOT (omitting it). Adding the promoter fragments to the figure would improve clarity.*
To clarify the design of the reporters, we have revised the text as follows: “(…) To test this idea, we generated three single-copy, integrated transcriptional reporters carrying a histone H2B sequence fused to an mNeon-Green (mNG) fluorescent protein sequence under the transcriptional control of the following dlg-1 upstream regions: (i) a full-length sequence (“FL” = Distal + uHOT + Proximal sequences), (ii) one spanning from the beginning of the HOT region to the dlg-1 CDS (“uHOT” = uHOT + Proximal sequences), and (iii) one starting at the end of the HOT region and ending at the dlg-1 CDS (“Short” = Proximal sequence) (Figure 1A-C). (…)”
This description clarifies which parts of the upstream region are included in each reporter and matches the schematics in Figure 1.
* Line 108. Re-work the phrase "increase majorly". Majorly increase would be better.*
We thank the reviewer for this suggestion. The verb is used here as an infinitive (“to increase majorly”), and in standard English the infinitive is usually not split. Therefore, we have kept the phrasing as it currently appears in the manuscript.
* Line 153-154. The deletion indicates that HOT2 and HOT2 were removed. Was one supposed to be HOT3?*
Thanks for spotting this typo. It has now been corrected.
* In the figure legends the number of animals scored and the number of biological repeats is missing.*
Added.
* Figure 1 title in the legend. Should read "main driver" not "man driver".*
Thanks for spotting this typo. It has now been corrected.
* The references need to be gone through carefully and cleaned up. There are numerous gene and species names that are not italicized. There are also extra elements added by the reference manager such as [Internet].*
Thanks for pointing it out. We used Zotero and the requested formatting from the journal of our choice. We will discuss with their team how to go through this issue.
-
-
-
www.biorxiv.org www.biorxiv.org
-
Note: This response was posted by the corresponding author to Review Commons. The content has not been altered except for formatting.
Learn more at Review Commons
Reply to the reviewers
__Reviewer #1 (Evidence, reproducibility and clarity (Required)): __
This study explores chromatin organization around trans-splicing acceptor sites (TASs) in the trypanosomatid parasites Trypanosoma cruzi, T. brucei and Leishmania major. By systematically re-analyzing MNase-seq and MNase-ChIP-seq datasets, the authors conclude that TASs are protected by an MNase-sensitive complex that is, at least in part, histone-based, and that single-copy and multi-copy genes display differential chromatin accessibility. Altogether, the data suggest a common chromatin landscape at TASs and imply that chromatin may modulate transcript maturation, adding a new regulatory layer to an unusual gene-expression system.
I value integrative studies of this kind and appreciate the careful, consistent data analysis the authors implemented to extract novel insights. That said, several aspects require clarification or revision before the conclusions can be robustly supported. My main concerns are listed below, organized by topic/result section.
TAS prediction * Why were TAS predictions derived only from insect-stage RNA-seq data? Restricting TAS calls to one life stage risks biasing predictions toward transcripts that are highly expressed in that stage and may reduce annotation accuracy for lowly expressed or stage-specific genes. Please justify this choice and, if possible, evaluate TAS robustness using additional transcriptomes or explicitly state the limitation.
TAS predictions derived only from insect-stage RNA-seq data because in a previous study it was shown that there are no significant differences between stages in the 5'UTR procesing in T. cruzi life stages (https://doi.org/10.3389/fgene.2020.00166) We are not testing an additional transcriptome here, because the robustness of the software was already probed in the original article were UTRme was described (Radio S, 2018 doi:10.3389/fgene.2018.00671).
Results - "There is a distinctive average nucleosome arrangement at the TASs in TriTryps": * You state that "In the case of L. major the samples are less digested." However, Supplementary Fig. S1 suggests that replicate 1 of L. major is less digested than the T. brucei samples, while replicate 2 of L. major looks similarly digested. Please clarify which replicates you reference and correct the statement if needed.
The reviewer has a good point. We made our statement based on the value of the maximum peak of the sequenced DNA molecules, which in general is a good indicative of the extension of the digestion achieved by the sample (Cole H, NAR, 2011).
As the reviewer correctly points, we should have also considered the length of the DNA molecules in each percentile. However, in this case both, T. brucei's and L major's samples were gel purified before sequencing and it is hard to know exactly what fragments were left behind in each case. Therefore, it is better not to over conclude on that regard.
We have now comment on this in the main manuscript, and we have clarified in the figure legends which data set we used in each case in the figure legends and in Table S1.
* It appears you plot one replicate in Fig. 1b and the other in Suppl. Fig. S2. Please indicate explicitly which replicate is in each plot. For T. brucei, the NDR upstream of the TAS is clearer in Suppl. Fig. S2 while the TAS protection is less prominent; based on your digestion argument, this should correspond to the more-digested replicate. Please confirm.
The replicates used for the construction of each figure are explicitly indicated in Table S1. Although we have detailed in the table the original publication, the project and accession number for each data set, the reviewer is correct that in this case it was still not completely clear to which length distribution heatmap was each sample associated with. To avoid this confusion, we have now added the accession number for each data set to the figure legends and also clarified in Table S1. Regarding the reviewer's comment on the correspondence between the observed TAS protection and the extent of samples digestion, he/she is correct that for a more digested sample we would expect a clearer NDR. In this case, the difference in the extent of digestion between these two samples is minor, as observed the length of the main peak in the length distribution histogram for sequenced DNA molecules is the same. These two samples GSM5363006, represented in Fig1 b, and GSM5363007, represented in S2, belong to the same original paper (Maree et al 2017), and both were gel purified before sequencing. Therefore, any difference between them could not only be the result of a minor difference in the digestion level achieved in each experiment but could be also biased by the fragments included or not during gel purification. Therefore, I would not over conclude about TAS protection from this comparison. We have now included a brief comment on this, in the figure discussion
* The protected region around the TAS appears centered on the TAS in T. brucei but upstream in L. major. This is an interesting difference. If it is technical (different digestion or TAS prediction offset), explain why; if likely biological, discuss possible mechanisms and implications.
We appreciate the reviewer suggestion. We cannot assure if it is due to technical or biological reasons, but there is evidence that L. major 's genome has a different dinucleotide content and it might have an impact on nucleosome assembly. We have now added a comment about this observation in the final discussion of the manuscript.
Additionally, we analyzed DRIP-seq data for L. major, recently published doi: 10.1038/s41467-025-56785-y, and we observed that the R-loop footprint co-localized with the MNase-protected region upstream of the TAS (new S5 Fig), suggesting that the shift is not related to the MNase-seq technique.
Results - "An MNase sensitive complex occupies the TASs in T. brucei": * The definition of "MNase activity" and the ordering of samples into Low/Intermediate/High digestion are unclear. Did you infer digestion levels from fragment distributions rather than from controlled experimental timepoints? In Suppl. Fig. S3a it is not obvious how "Low digestion" was defined; that sample's fragment distribution appears intermediate. Please provide objective metrics (e.g., median fragment length, fraction 120-180 bp) used to classify digestion levels.
As the reviewer suggests, the ideal experiment would be to perform a time course of MNase reaction with all the samples in parallel, or to work with a fixed time point adding increasing amounts of MNase. However, even when making controlled experimental timepoints, you need to check the length distribution histogram of sequenced DNA molecules to be sure which level of digestion you have achieved.
In this particular case, we used public available data sets to make this analysis. We made an arbitrary definition of low, intermediate and high level of digestion, not as an absolute level of digestion, but as a comparative output among the tested samples. We based our definition on the comparison of __the main peak in length distribution heatmaps because this parameter is the best metric to estimate the level of digestion of a given sample. It represents the percentage of the total DNA sequenced that contains the predominant length in the sample tested. __Hence, we considered:
low digestion: when the main peak is longer than the expected protection for a nucleosome (longer than 150 bp). We expect this sample to contain additional longer bands that correspond to less digested material.
intermediate digestion, when the main peak is the expected for the nucleosome core-protection (˜146-150bp).
high digestion, when the main peak is shorter than that (shorter than 146 bp). This case, is normally accompanied by a bigger dispersion in fragment sizes.
To do this analysis, we chose samples that render different MNase protection of the TAS when plotting all the sequenced DNA molecules relative to this point and we used this protection as a predictor of the extent of sample digestion (Figure 2). To corroborate our hypothesis, that the degree of TAS protection was indeed related to the extent of the MNase digestion of a given sample, we looked at the length distribution histogram of the sequenced DNA molecules in each case. It is the best measurement of the extent of the digestion achieved, especially, when sequencing the whole sample without any gel purification and representing all the reads in the analysis as we did. The only caveat is with the sample called "intermediate digestion 1" that belongs to the original work of Mareé 2017, since only this data set was gel purified. To avoid this problem, we decided to remove this data from figures 2 and S3. In summary, the 3 remaining samples comes from the same lab, and belong to the same publication (Mareé 2022). These sample are the inputs of native MNase ChIp-seq, obtain the same way, totally comparable among each other.
* Several fragment distributions show a sharp cutoff at ~100-125 bp. Was this due to gel purification or bioinformatic filtering? State this clearly in Methods. If gel purification occurred, that can explain why some datasets preserve the MNase-sensitive region.
The sharp cutoff is neither due to gel purification or bioinformatic filtering, it is just due to the length of the paired-end read used in each case. In earlier works the most common was to sequence only 50bp, with the improvement of technologies it went up to 75,100 or 125 bp. We have now clarified in Table S1 the length of the paired-reads used in each case when possible.
* Please reconcile cases where samples labeled as more-digested contain a larger proportion of >200 bp fragments than supposedly less-digested samples; this ordering affects the inference that digestion level determines the loss/preservation of TAS protection. Based on the distributions I see, "Intermediate digestion 1" appears most consistent with an expected MNase curve - please confirm and correct the manuscript accordingly.
As explained above, it's a common observation in MNase digestion of chromatin that more extensive digestion can still result in a broad range of fragment sizes, including some longer fragments. This seemingly counter-intuitive result is primarily due to the non-uniform accessibility of chromatin and the sequence preference of the MNase enzyme, which has a preference for AT reach sequences.
The rationale of this is as follows: when you digest chromatin with MNase and the objective is to map nucleosomes genome-wide, the ideal situation would be to get the whole material contained in the mononucleosome band. Given that MNase is less efficient to digest protected DNA but, if the reaction proceeds further, it always ends up destroying part of it, the result is always far from perfect. The better situation we can get, is to obtain samples were ˜80% of the material is contained in the mononucloesome band. __And here comes the main point: __even in the best scenario, you always get some additional longer bands, such as those for di or tri nucleosomes. If you keep digesting, you will get less than 80 % in the nucleosome band and, those remaining DNA fragments that use to contain di and tri nucleosomes start getting digested as well, originating a bigger dispersion in fragments sizes. How do we explain persistence of Long Fragments? The longest fragments (di-, tri-nucleosomes) that persist in a highly digested sample are the ones that were originally most highly protected by proteins or higher-order structure, or by containing a poor AT sequence content, making their linker DNA extremely resistant to initial cleavage. Once the majority of the genome is fragmented, these few resistant longer fragments become a more visible component of the remaining population, contributing to a broader size dispersion. Hence, you end up observing a bigger dispersion in length distributions in the final material. Bottom line, it is not a good practice to work with under or over digested samples. Our main point, is to emphasize that especially when comparing samples, it important to compare those with comparable levels of digestion. Otherwise, a different sampling of the genome will be represented in the remaining sequenced DNA.
Results - "The MNase sensitive complexes protecting the TASs in T. brucei and T. cruzi are at least partly composed of histones": * The evidence that histones are part of the MNase-sensitive complex relies on H3 MNase-ChIP signal in subnucleosomal fragment bins. This seems to conflict with the observation (Fig. 1) that fragments protecting TASs are often nucleosome-sized. Please reconcile these points: are H3 signals confined to subnucleosomal fragments flanking the TAS while the TAS itself is depleted of H3? Provide plots that compare MNase-seq and H3 ChIP signals stratified by consistent fragment-size bins to clarify this.
What we learned from other eukaryotic organisms that were deeply studied, such as yeast, is that NDRs are normally generated at regulatory points in the genome. In this sense, yeast tRNA genes have a complex with a bootprint smaller than a nucleosome formed by TFIIIC-TFIIB (Nagarajavel, doi: 10.1093/nar/gkt611). On the other hand, many promotor regions have an MNase-sensitive complex with a nucleosome-size footprint, but it does not contain histones (Chereji, et al 2017, doi:10.1016/j.molcel.2016.12.009). The reviewer is right that from Figure 1 and S2 we could observe that the footprint of whatever occupies the TAS region, especially in T. brucei, is nucleosome-size. However, it only shows the size, but it doesn't prove the nature of its components. Nevertheless, those are only MNase-seq data sets. Since it does not include a precipitation with specific antibodies, we cannot confirm the protecting complex is made up by histones. In parallel, a complementary study by Wedel 2017, from Siegel's lab, shows that using a properly digested sample and further immunoprecipitating with a-H3 antibody, the TAS is not protected by nucleosomes at least not when analyzing nucleosome size-DNA molecules. Besides, Briggs et. al 2018 (doi: 10.1093/nar/gky928) showed that at least at intergenic regions H3 occupancy goes down while R-loops accumulation increases. We have now added a new figure 4 replotting R-loops and MNase-ChIP-seq for H3 relative to our predicted TAS showing this anti-correlation and how it partly correlates with MNase protection as well. As a control we show that Rpb9 trends resembles H3 as Siegel's lab have shown in Wedel 2018. Moreover, we analyzed redate from a recently published paper (doi: 10.1038/s41467-025-56785-y) added a new supplemental figure 5 showing that a similar correlation between MNase protection and R-loop footprint occurs in L. major (S5 Fig).
* Please indicate which datasets are used for each panel in Suppl. Fig. S4 (e.g., Wedel et al., Maree et al.), and avoid calling data from different labs "replicates" unless they are true replicates.
In most of our analysis we used real replicated experiments. Such is the case MNase-seq data used in Figure 1, with the corresponding replicate experiments used in Figure S2; T. cruzi MNase-ChIP-seq data used in Figure 3b and 4a with the respective replicate used in Figures S4 and S5 (now S6 in the revised manuscript). The only case in which we used experiments coming from two different laboratories, is in the case of MNase-ChIP-seq for H3 from T. brucei. Unfortunately, there are only two public data sets coming each of them from different laboratories. The samples used in Fig 3 (from Siegel's lab) whether the IP from H3 represented in S4 and S5 (S6 n the updated version) comes from another lab (Patterton's). To be more rigorous, we now call them data 1 and 2 when comparing these particular case.
The reviewer is right that in this particular case one is native chromatin (Pattertons') while the other one is crosslinked (Siegel's). We have now clarified it in the main text that unfortunately we do not count on a replicate but even under both condition the result remains the same, and this is compatible with my own experience, were crosslinking does not affect the global nucleosome patterns (compared nucleosome organization from crosslinked chromatin MNAse-seq inputs Chereji, Mol Cell, 2017 doi: 10.1016/j.molcel.2016.12.009 and native MNase-seq from Ocampo, NAR, 2016 doi: 10.1093/nar/gkw068).
* Several datasets show a sharp lower bound on fragment size in the subnucleosomal range (e.g., ~80-100 bp). Is this a filtering artifact or a gel-size selection? Clarify in Methods and, if this is an artifact, consider replotting after removing the cutoff.
We have only filtered adapter dimmer or overrepresented sequences when needed. In Figures 2 and S3 we represented all the sequenced reads. In other figures when we sort fragments sizes in silico, such as nucleosome range, dinucleosome or subnucleosome size, we make a note in the figure legends. What the reviewer points is related to the length of the sequence DNA fragment in each experiment. As we explained above, the older data-sets were performed with 50 bp paired-end reads, the newer ones are 75, 100 or 125bp. This is information is now clarified in Table S1.
__Results - "The TASs of single and multi-copy genes are differentially protected by nucleosomes": __
__ __* Please include T. brucei RNA-seq data in Suppl. Fig. S5b as you did for T. cruzi.
We have shown chromatin organization for T. brucei in previous S5b to illustrate that there is a similar trend. Unfortunately, we did not get a robust list of multi-copy genes for T. brucei as we did get for T. cruzi, therefore we do not want to over conclude showing the RNA-seq for these subsets of genes. The limitation is related to the fact that UTRme restrict the search and is extremely strict when calling sites at repetitive regions. Additionally, attending to the request of one reviewer we have now changed the UTR predictions for T. brucei using a different RNA-seq data set from Lister 427(detail in method section). Given that with the new predictions it was even harder to obtain the list of multicopy genes for T. brucei, we decided to remove that figure in the updated version of the manuscript.
* Discuss how low or absent expression of multigene families affects TAS annotation (which relies on RNA-seq) and whether annotation inaccuracies could bias the observed chromatin differences.
The mapping of occurrence and annotations that belong to repetitive regions has great complexity. UTRme is specially designed to avoid overcalling those sites. In other words, there is a chance that we could be underestimating the number of predicted TASs at multi-copy genes. Regarding the impact on chromatin analysis, we cannot rule out that it might have an impact, but the observation favors our conclusion, since even when some TASs at multi-copy genes can remain elusive, we observe more nucleosome density at those places.
* The statement that multi-copy genes show an "oscillation" between AT and GC dinucleotides is not clearly supported: the multi-copy average appears noisier and is based on fewer loci. Please tone down this claim or provide statistical support that the pattern is periodic rather than noisy.
We have fixed this now in the preliminary revised version
* How were multi-copy genes defined in T. brucei? Include the classification method in Methods.
This classification was done the same way it was explained for T. cruzi. However, decided to remove the supplemental figure that included this sorting.
Genomes and annotations: * If transcriptomic data for the Y strain was used for T. cruzi, please explain why a Y strain genome was not used (e.g., Wang et al. 2021 GCA_015033655.1), or justify the choice. For T. brucei, consider the more recent Lister 427 assembly (Tb427_2018) from TriTrypDB. Use strain-matched genomes and transcriptomes when possible, or discuss limitations.
The most appropriate way to analyze high throughput data, is to aline it to the same genome were the experiments were conducted. This was clearly illustrated in a previous publication from our group were we explained how should be analyzed data from the hybrid CL Brener strain. A common practice in the past was to use only Esmeraldo-like genome for simplicity, but this resulted in output artifacts. Therefore, we aligned it to CL Brener genome, and then focused the main analysis on the Esmeraldo haplotype (Beati Plos ONE, 2023). Ideally, we should have counted on transcriptomic data for the same strain (CL Brener or Esmeraldo). Since this was not the case at that moment, we used data from Y strain that belongs to the same DTU with Esmeraldo.
In the case of T. brucei, when we started our analysis and the software code for UTRme was written, the previous version of the genome was available. Upon 2018 version came up, we checked chromatin parameters and observed that it did not change the main observations. Therefore, we continue working with our previous setups.
Reproducibility and broader integration: * Please share the full analysis pipeline (ideally on GitHub/Zenodo) so the results are reproducible from raw reads to plots.
We are preparing a full pipeline in GitHub. We will make it available before manuscript full revision
* As an optional but helpful expansion, consider including additional datasets (other life stages, BSF MNase-seq, ATAC-seq, DRIP-seq) where available to strengthen comparative claims.
We are now including a new figure 4 and a supplemental figure 5 including DRIP-seq and Rp9 ChIP-seq for T. brucei (revised Fig 4) and DRIP-seq for L. major (S5 Fig). Additionally, we added FAIRE-seq data to previous Fig 4 now Fig 5 (revised Fig 5C).
We are analyzing ATAC-seq data for T. brucei.
Regarding BSF MNase-seq, the original article by Mareé 2017 claims that there is not significant difference for average chromatin organization between the two life forms; therefore, is not worth including that analysis.
Optional analyses that would strengthen the study: * Stratify single-copy genes by expression (high / medium / low) and examine average nucleosome occupancy at TASs for each group; a correlation between expression and NDR depth would strengthen the functional link to maturation.
We have now included a panel in suplemental figure 5 (now revised S6), showing the concordance for chromatin organization of stratified genes by RNA-seq levels relative to TAS.
__Minor / editorial comments: __ * In the Introduction, the sentence "transcription is initiated from dispersed promoters and in general they coincide with divergent strand switch regions" should be qualified: such initiation sites also include single transcription start regions.
We have clarified this in the preliminary revised version
* Define the dotted line in length distribution plots (if it is not the median, please clarify) and consider placing it at 147 bp across plots to ease comparison.
The dotted line is just to indicate where the maximum peak is located. It is now clarified in figure legends.
* In Suppl. Fig. 4b "Replicate2" the x-axis ticks are misaligned with labels - please fix.
We have now fixed the figure. Thanks for noticing this mistake.
* Typo in the Introduction: "remodellingremodeling" → "remodeling
Thanks for noticing this mistake, it is fixed in the current version of the manuscript
**Referee cross-commenting** Comment 1: I think Reviewer #2 and Reviewer #3 missed that they authors of this manuscript do cite and consider the results from Wedel at al. 2017. They even re-analysed their data (e.g. Figure 3a). I second Reviewer #2 comment indicating that the inclusion of a schematic figure to help readers visualize and better understand the findings would be an important addition.
Comment 2: I agree with Reviewer #3 that the use of different MNase digestion procedures in the different datasets have to be considered. On the other hand, I don't think there is a problem with figure 1 showing an MNase-protected TAS for T. brucei as it is based on MNase-seq data and reproduces the reported results (Maree et al. 2017). What the Siegel lab did in Wedel et al. 2017 was MNase-ChIPseq of H3 showing nucleosome depletion at TAS, but both results are not necessary contradictory: There could still be something else (which does not contain H3) sitting on the TAS protecting it from MNase digestion.
Reviewer #1 (Significance (Required)):
This study provides a systematic comparative analysis of chromatin landscapes at trans-splicing acceptor sites (TASs) in trypanosomatids, an area that has been relatively underexplored. By re-analyzing and harmonizing existing MNase-seq and MNase-ChIP-seq datasets, the authors highlight conserved and divergent features of nucleosome occupancy around TASs and propose that chromatin contributes to the fidelity of transcript maturation. The significance lies in three aspects: 1. Conceptual advance: It broadens our understanding of gene regulation in organisms where transcription initiation is unusual and largely constitutive, suggesting that chromatin can still modulate post-transcriptional processes such as trans-splicing. 2. Integrative perspective: Bringing together data from T. cruzi, T. brucei and L. major provides a comparative framework that may inspire further mechanistic studies across kinetoplastids. 3. Hypothesis generation: The findings open testable avenues about the role of chromatin in coordinating transcript maturation, the contribution of DNA sequence composition, and potential interactions with R-loops or RNA-binding proteins. Researchers in parasitology, chromatin biology, and RNA processing will find it a useful resource and a stimulus for targeted experimental follow-up.
My expertise is in gene regulation in eukaryotic parasites, with a focus on bioinformatic analysis of high-throughput sequencing data
__Reviewer #2 (Evidence, reproducibility and clarity (Required)): __
Siri et al. perform a comparative analysis using publicly available MNase-seq data from three trypanosomatids (T. brucei, T. cruzi, and Leishmania), showing that a similar chromatin profile is observed at TAS (trans-splicing acceptor site) regions. The original studies had already demonstrated that the nucleosome profile at TAS differs from the rest of the genome; however, this work fills an important gap in the literature by providing the most reliable cross-species comparison of nucleosome profiles among the tritryps. To achieve this, the authors applied the same computational analysis pipeline and carefully evaluated MNase digestion levels, which are known to influence nucleosome profiling outcomes.
In my view, the main conclusion is that the profiles are indeed similar-even when comparing T. brucei and T. cruzi. This was not clear in previous studies (and even appeared contradictory, reporting nucleosome depletion versus enrichment) largely due to differences in chromatin digestion across these organisms. The manuscript could be improved with some clarifications and adjustments:
- The authors state from the beginning that available MNase data indicate altered nucleosome occupancy around the TAS. However, they could also emphasize that the conclusions across the different trypanosomatids are inconsistent and even contradictory: NDR in T. cruzi versus protection-in different locations-in T. brucei and Leishmania.
We start our manuscript by referring to the first MNase-seq data sets publicly available for each TriTryp and we point that one of the main observations, in each of them, is the occurrence of a change in nucleosome density or occupancy at intergenic regions. In T. cruzi, in a previous publication from our group, we stablished that this intergenic drop in nucleosome density occurs near the trans-splicing acceptor site. In this work, we extend our study to the other members of TriTryps: T. brucei and L. major.
In T. brucei the papers from Patterton's lab and Siegel's lab came out almost simultaneously in 2017. Hence, they do not comment on each other's work. The first one claims the presence of a well-positioned nucleosome at the TAS by using MNase-seq, while the second one, shows an NDR at the TAS by using MNase-ChIP-seq. However, we do not think they are contradictory, or they have inconsistency. We brought them together along the manuscript because we think these works can provide complementary information.
On one hand, we infer data from Pattertons lab is slightly less digested than the sample from Siegel's lab. Therefore, we discuss that this moderate digestion must be the reason why they managed to detect an MNase protecting complex sitting at the TAS (Figure 1). On the other hand, Sigel's lab includes an additional step by performing MNase-ChIP-seq, showing that when analyzing nucleosome size fragments, histones are not detected at the TAS. Here, we go further in this analysis on figure 3, showing that only when looking at subnucleosome-size fragments, we can detect histone H3. And this is also true for T. cruzi.
By integrating every analysis in this work and the previous ones, we propose that TASs are protected by an MNase-sensitive complex (proved in Figure 2). This complex most likely is only partly formed by histones, since only when analyzing sub-nucleosomes size DNA molecules we can detect histone H3 (Figure 3). To be sure that the complex is not entirely made up by histones, future studies should perform an MNse-ChIP-seq with less digested samples. However, it was previously shown that R-loops are enriched at those intergenic NDRs (Briggs, 2018 doi: 10.1093/nar/gky928) and that R-loops have plenty of interacting proteins (Girasol, 2023 10.1093/nar/gkad836). Therefore, most likely, this MNase-sensitive complexed have a hybrid nature made up by H3 and some other regulatory molecules, possibly involved in trans-splicing. We have now added a new figure 4 showing R-loop co-localization with the NDR.
Regarding the comparison between different organisms, after explaining the sensitivity to MNase of the TAS protecting complex, we discuss that when comparing equally digested samples T. cruzi and T. brucei display a similar chromatin landscape with a mild NDR at the TAS (See T. cruzi represented in Figure 1 compared to T. brucei represented in Intermediate digestion 2 in Figure 2, intermediate digestion in the revised manuscript). Unfortunately, we cannot make a good comparison with L. major, since we do not count on a similar level of digestion. However, by analyzing a recently published DRIP-seq data-set for L. major we show that R-loop signal co localize with MNase-protection in a similar way (new S5 Fig).
Another point that requires clarification concerns what the authors mean in the introduction and discussion when they write that trypanosomes have "...poorly organized chromatin with nucleosomes that are not strikingly positioned or phased." On the other hand, they also cite evidence of organization: "...well-positioned nucleosome at the spliced-out region.. in Leishmania (ref 34)"; "...a well-positioned nucleosome at the TASs for internal genes (ref37)"; "...a nucleosome depletion was observed upstream of every gene (ref 35)." Aren't these examples of organized chromatin with at least a few phased nucleosomes? In addition, in ref 37, figure 4 shows at least two (possibly three to four) nucleosomes that appear phased. In my opinion, the authors should first define more precisely what they mean by "poorly organized chromatin" and clarify that this interpretation does not contradict the findings highlighted in the cited literature.
For a better understanding of nucleosome positioning and phasing I recommend the review: Clark 2010 doi:10.1080/073911010010524945, Figure 4. Briefly, in a cell population there are different alternative positions that a given nucleosome can adopt. However, some are more favorable. When talking about favorable positions, we refer to the coordinates in the genome that are most likely covered by a nucleosome and are predominant in the cell population. Additionally, nucleosomes could be phased or not. This refers not only the position in the genome, but to the distance relative to a given point. In yeast, or in highly transcribed genes of more complex eukaryotes, nucleosomes are regularly spaced and phased relative to the transcription start site (TSS) or to the +1 nucleosome (Ocampo, NAR, 2016, doi:10.1093/nar/gkw068). In trypanosomes, nucleosomes have some regular distribution when making a browser inspection but, given that they are not properly phased with respect to any point, it is almost impossible to make a spacing estimation from paired-end data. This is also consistent with a chromatin that is transcribed in an almost constitutive manner.
As the reviewer mention, we do site evidence of organization. We think the original observations are correct, but we do not fully agree with some of the original statements. In this manuscript our aim is to take the best we learned from their original works and to make a constructive contribution adding to the original discussions. In this regard, in trypanosomes there are some conserved patterns in the chromatin landscape, but their nucleosomes are far from being well-positioned or phased. For a better understanding, compare the variations observed in the y axis when representing av. nucleosome occupancy in yeast with those observed in trypanosomes and you will see that the troughs and peaks are much more prominent in yeast than the ones observed in any TryTryp member.
Following the reviewer's suggestion we have now clarified this in the main text.
The paper would also benefit from the inclusion of a schematic figure to help readers visualize and better understand the findings. What is the biological impact of having nucleosomes, di-nucleosomes, or sub-nucleosomes at TAS? This is not obvious to readers outside the chromatin field. For example, the following statement is not intuitive: "We observed that, when analyzing nucleosome-size (120-180 bp) DNA molecules or longer fragments (180-300 bp), the TASs of either T. cruzi or T. brucei are mostly nucleosome-depleted. However, when representing fragments smaller than a nucleosome-size (50-120 bp) some histone protection is unmasked (Fig. 3 and Fig. S4). This observation suggests that the MNase sensitive complex sitting at the TASs is at least partly composed of histones." Please clarify.
We appreciate the reviewer's suggestion to make a schematic figure. We have now added a new Figure 6.
Regarding the biological impact of having mono, di or subnucleosome fragments, it is important to unveil the fragment size of the protected DNA to infer the nature of the protecting complex. In the case of tRNA genes in yeast, at pol III promoters they found footprints smaller than a nucleosome size that ended up being TFIIB-TFIIC (Nagarajavel, doi: 10.1093/nar/gkt611). Therefore, detecting something smaller than a nucleosome might suggest the binding of trans-acting factors different than histones or involving histones in a mixed complex. These mixed complexes are also observed, and that is the case of the centromeric nucleosome which has a very peculiar composition (Ocampo and Clark, Cells Reports, 2015). On the other hand, if instead we detect bigger fragments, it could be indicative of the presence of bigger protecting molecules or that those regions are part of higher order chromatin organization still inaccessible for MNase linker digestions.
Here we show on 2Dplots, that complex or components protecting the TAS have nucleosome size, but we cannot assure they are entirely made up by histones, since, only when looking at subnucleosome-size fragments, we are able to detect histone H3. We have now added part of this explanation to the discussion.
By integrating every analysis in this work and the previous ones, we propose that the TAS is protected by an MNase-sensitive complex (Figure 2). This complex most likely is only partly formed by histones, since only when analyzing sub-nucleosomes size DNA molecules we can detect histone H3 (Figure 3). As explained above, to be sure that the complex is not entirely made up by histones, future studies should perform an MNse-ChIP-seq with less digested samples. However, it was previously shown that R-loops are enriched at those intergenic NDRs (Briggs 2018) and that R-loops have plenty of interacting proteins (Girasol, 2023). Therefore, most likely, this MNase-sensitive complexed have a hybrid nature made up by H3 and some other regulatory molecules. We have now added a new figure 4 showing R-loop partial co-localization with MNase protection.
Some references are missing or incorrect:
we will make a thorough revision
"In trypanosomes, there are no canonical promoter regions." - please check Cordon-Obras et al. (Navarro's group). Thank you for the appropiate suggestion.
Thank you for the appropriate suggestion. We have now added this reference
Please, cite the study by Wedel et al. (Siegel's group), which also performed MNase-seq analysis in T. brucei.
We understand that reviewer number 2# missed that we cited this reference and that we did used the raw data from the manuscript of Wedel et. al 2017 form Siegel's group. We used the MNase-ChIP-seq data set of histone H3 in our analysis for Figures 3, S4 and S6 (in the revised version), also detailed in table S1. To be even more explicit, we have now included the accession number of each data set in the figure legends.
Figure-specific comments: Fig. S3: Why does the number of larger fragments increase with greater MNase digestion? Shouldn't the opposite be expected?
This a good observation. As we also explained to reviewer#1:
It's a common observation in MNase digestion of chromatin that more extensive digestion can still result in a broad range of fragment sizes, including some longer fragments. This seemingly counter-intuitive result is primarily due to the non-uniform accessibility of chromatin and the sequence preference of the MNase enzyme.
The rationale of this is as follows: when you digest chromatin with MNase and the objective is to map nucleosomes genome-wide, the ideal situation would get the whole material contained in the mononucleosome band. Given that MNase is less efficient to digest protected DNA but, if the reaction proceeds further, it always ends up destroying part of it, the result is always far from perfect. The better situation we can get, is to obtain samples were ˜80% of the material is contained in the mononucloesome band. __And here comes the main point: __even in the best scenario, you always have some additional longer bands, such as those for di or tri nucleosomes. If you keep digesting, you will get less than 80 % in the nucleosome band and, those remaining DNA fragments that use to contain di and tri nucleosomes start getting digested as well originating a bigger dispersion in fragments sizes. How do we explain persistence of Long Fragments? The longest fragments (di-, tri-nucleosomes) that persist in a highly digested sample are the ones that were originally most highly protected by proteins or higher-order structure, making their linker DNA extremely resistant to initial cleavage. Once most of the genome is fragmented, these few resistant longer fragments become a more visible component of the remaining population, contributing to a broader size dispersion. Hence, there you end up having a bigger dispersion in length distributions in the final material. Bottom line, it is not a good practice to work with under or overdirected samples. Our main point is to emphasize that especially when comparing samples, it important to compare those with comparable levels of digestion. Otherwise, a different sampling of the genome will be represented in the remaining sequenced DNA.
Minor points:
There are several typos throughout the manuscript.
Thanks for the observation. We will check carefully.
Methods: "Dinucelotide frecuency calculation."
We will add a code in GitHub
Reviewer #2 (Significance (Required)):
In my view, the main conclusion is that the profiles are indeed similar-even when comparing T. brucei and T. cruzi. This was not clear in previous studies (and even appeared contradictory, reporting nucleosome depletion versus enrichment) largely due to differences in chromatin digestion across these organisms. Audience: basic science and specialized readers.
Expertise: epigenetics and gene expression in trypanosomatids.
__Reviewer #3 (Evidence, reproducibility and clarity (Required)): __
The authors analysed publicly accessible MNase-seq data in TriTryps parasites, focusing on the chromatin structure around trans-splicing acceptor sites (TASs), which are vital for processing gene transcripts. They describe a mild nucleosome depletion at the TAS of T. cruzi and L. major, whereas a histone-containing complex protects the TASs of T. brucei. In the subsequent analysis of T. brucei, they suggest that a Mnase-sensitive complex is localised at the TASs. For single-copy versus multi-copy genes, the authors show different di-nucleotide patterns and chromatin structures. Accordingly, they propose this difference could be a novel mechanism to ensure the accuracy of trans-splicing in these parasites.
Before providing an in- depth review of the manuscript, I note that some missing information would have helped in assessing the study more thoroughly; however, in the light of the available information, I provide the following comments for consideration.
The numbering of the figures, including the figure legends, is missing in the PDF file. This is essential for assessing the provided information.
We apologized for not including the figure numbers in the main text, although they are located in the right place when called in the text. The omission was unwillingly made when figure legends were moved to the bottom of the main text. This is now fixed in the updated version of the manuscript.
The publicly available Mnase- seq data are manyfold, with multiple datasets available for T. cruzi, for example. It is unclear from the manuscript which dataset was used for which figure. This must be clarified.
This was detailed in Table S1. We have now replaced the table by an improved version, and we have also included the accession number of each data set used in the figure legends.
Why do the authors start in figure 1 with the description of an MNase- protected TAS for T.brucei, given that it has been clearly shown by the Siegel lab that there is a nucleosome depletion similar to other parasites?
We did not want to ignore the paper from Patterton's lab because it was the first one to map nucleosomes genome-wide in T. brucei and the main finding of that paper claimed the existence of a well-positioned nucleosome at intergenic regions, what we though constitutes a point worth to be discussed. While Patterton's work use MNase-seq from gel-purified samples and provides replicated experiments sequenced in really good depth; Siegel's lab uses MNase-ChIP-seq of histone H3 but performs only one experiment and its input was not sequenced. So, each work has its own caveats and provides different information that together contributes to make a more comprehensive study. We think that bringing up both data sets to the discussion, as we have done in Figures 1 and 3, helps us and the community working in the field to enrich the discussion.
If the authors re- analyse the data, they should compare their pipeline to those used in the other studies, highlighting differences and potential improvements.
We are working on this point. We will provide a more detail description in the final revision.
Since many figures resemble those in already published studies, there seems little reason to repeat and compare without a detailed comparison of the pipelines and their differences.
Following the reviewer advice, we are now working on highlighting the main differences that justify analyzing the data the way we did and will be added in the finally revised method section.
At a first glance, some of the figures might look similar when looking at the original manuscripts comparing with ours. However, with a careful and detailed reading of our manuscripts you can notice that we have added several analyses that allow to unveil information that was not disclosed before.
First, we perform a systematic comparison analyzing every data set the same way from beginning to end, being the main difference with previous studies the thorough and precise prediction of TAS for the three organisms. Second, we represent the average chromatin organization relative to those predicted TASs for TriTryps and discuss their global patterns. Third, by representing the average chromatin into heatmaps, we show for the very first time, that those average nucleosome landscape are not just an average, they keep a similar organization in most of the genome. These was not done in any of the previous manuscripts except for our own (Beati, PLOS One 2023). Additionally, we introduce the discussion of how the extension of MNase reaction can affect the output of these experiments and we show 2D-plots and length distribution heatmaps to discuss this point (a point completely ignored in all the chromatin literature for trypanosomes). Furthermore, we made a far-reaching analysis by considering the contributions of each publish work even when addressed by different techniques. Finally, we discuss our findings in the context of a topic of current interest in the field, such as TriTryp's genome compartmentalization.
Several previous Mnase- seq analysis studies addressing chromatin accessibility emphasized the importance of using varying degrees of chromatin digestion, from low to high digestion (30496478, 38959309, 27151365).
The reviewer is correct, and this point is exactly what we intended to illustrate in figure number 2. We appreciate he/she suggests these references that we are now citing in the final discussion. Just to clarify, using varying degrees of chromatin digestion is useful to make conclusions about a given organism but when comparing samples, strains, histone marks, etc. It is extremely important to do it upon selection of similar digested samples.
No information on the extent of DNA hydrolysis is provided in the original Mnase- seq studies. This key information can not be inferred from the length distribution of the sequenced reads.
The reviewer is correct that "No information on the extent of DNA hydrolysis is provided in the original Mnase-seq studies" and this is another reason why our analysis is so important to be published and discussed by the scientific community working in trypanosomes. We disagree with the reviewer in the second statement, since the level of digestion of a sequenced sample is actually tested by representing the length distribution of the total DNA sequenced. It is true that before sequencing you can, and should, check the level of digestion of the purified samples in an agarose gel and/or in a bioanalyzer. It could be also tested after library preparation, but before sequencing, expecting to observe the samples sizes incremented in size by the addition of the library adapters. But, the final test of success when working with MNase digested samples is to analyze length of DNA molecules by representing the histograms with length distribution of the sequenced DNA molecules. Remarkably, on occasions different samples might look very similar when run in a gel, but they render different length distribution histograms and this is because the nucleosome core could be intact but they might have suffered a differential trimming of the linker DNA associated to it or even be chewed inside (see Cole Hope 2011, section 5.2, doi: 10.1016/B978-0-12-391938-0.00006-9, for a detailed explanation).
As the input material are selected, in part gel- purified mono- nucleosomal DNA bands. Furthermore the datasets are not directly comparable, as some use native MNase, while others employ MNase after crosslinking; some involve short digestion times at 37 {degree sign} C, while others involve longer digestion at lower temperatures. Combining these datasets to support the idea of an MNase- sensitive complex at the TAS of T. brucei therefore may not be appropriate, and additional experiments using consistent methodologies would strengthen the study's conclusions.
In my opinion, describing an MNase- sensitive complex based solely on these data is not feasible. It requires specifically designed experiments using a consistent method and well- defined MNase digestion kinetics.
As the reviewer suggests, the ideal experiment would be to perform a time course of MNase reaction with all the samples in parallel, or to work with a fix time point adding increasing amounts of MNase. However, the information obtained from the detail analysis of the length distribution histogram of sequenced DNA molecules the best test of the real outcome. In fact, those samples with different digestion levels were probably not generated on purpose.
The only data sets that were gel purified are those from Mareé 2017 (Patterton's lab), used in Figures 1, S1 and S2 and those from L. major shown in Fig 1. It was a common practice during those years, then we learned that is not necessary to gel purify, since we can sort fragment sizes later in silico when needed.
As we explained to reviewer #1, to avoid this conflict, we decided to remove this data from figures 2 and S3. In summary, the 3 remaining samples comes from the same lab, and belong to the same publication (Mareé 2022). These sample are the inputs of native MNase ChIp-seq, obtain the same way, totally comparable among each other.
Reviewer #3 (Significance (Required)):
Due to the lack of controlled MNase digestion, use of heterogeneous datasets, and absence of benchmarking against previous studies, the conclusions regarding MNase-sensitive complexes and their functional significance remain speculative. With standardized MNase digestion and clearly annotated datasets, this study could provide a valuable contribution to understanding chromatin regulation in TriTryps parasites.
As we have explained in the previous point our conclusions are valid since we do not compare in any figure samples coming from different treatments. The only exception to this comment could be in figure 3 when talking about MNase-ChIP-seq. We have now added a clear and explicit comment in the section and the discussion that despite having subtle differences in experimental procedures we arrive to the same results. This is the case for T. cruzi IP, run from crosslinked chromatin, compared to T. brucei's IP, run from native chromatin.
Along the years it was observed in the chromatin field that nucleosomes are so tightly bound to DNA that crosslinking is not necessary. However, it is still a common practice specially when performing IPs. In our own hands, we did not observe any difference at the global level neither in T. cruzi (unpublished) nor in my previous work with yeast (compared nucleosome organization from crosslinked chromatin MNAse-seq inputs Chereji, Mol Cell, 2017 doi:10.1016/j.molcel.2016.12.009 and native MNase-seq from Ocampo, NAR, 2016 doi: 10.1093/nar/gkw068).
-
-
www.carlboettiger.info www.carlboettiger.info
-
Welcome to my Lab Notebook - Reloaded Welcome to my lab notebook, version 3.0. My original open lab notebooks began on the wiki platform OpenWetWare, moved to a personally hosted Wordpress platform, and now run on a Jekyll-powered platform (site-config), but the basic idea remains the same. For completeness, earlier entries from both platforms have been migrated here. Quoting from my original introduction to the Wordpress notebook: Disclaimer: Not a Blog Welcome to my open lab notebook. This is the active, permanent record of my scientific research, standing in place of the traditional paper bound lab notebook. The notebook is primarily a tool for me to do science, not communicate it. I write my entries with the hope that they are intelligible to my future self; and maybe my collaborators and experts in my field. Only the occasional entry will be written for a more general audience. […] In these pages you will find not only thoughts and ideas, but references to the literature I read, the codes or manuscripts I write, derivations I scribble and graphs I create and mistakes I make. Why an open notebook? Is it working? My original introduction to the notebook from November 2010 dodged this question by suggesting the exercise was merely an experiment to see if any of the purported benefits or supposed risks were well-founded. Nearly three years in, can I draw any conclusions from this open notebook experiment? In that time, the notebook has seen six projects go from conception to publication, and a seventh founder on a null result (see #tribolium). Several more projects continue to unfold. I have often worked on several projects simultaneously, and some projects branch off while others merge, making it difficult to capture all the posts associated with a single paper into a single tag or category. Of course not all ideas make it into the paper, but they remain captured in the notebook. I often return to my earlier posts for my own reference, and frequently pass links to particular entries to collaborators or other colleagues. On occasion I have pointed reviewers of my papers to certain entries discussing why we did y instead of x, and so forth. Both close colleagues and researchers I’ve never met have emailed me to follow up on something they had read in my notebook. This evidence suggests that the practice of open notebook science can faciliate both the performance and dissemination of research while remaining compatible and even synergistic with academic publishing. I am both proud and nervous to know of a half dozen other researchers who have credited me for inspiring them to adopt open or partially open lab notebooks online. I am particularly grateful for the examples, interactions, and ideas from established practitioners of open notebook science in other fields. My collaborators have been largely been somewhere between favorable and agnostic towards the idea, with the occasional request for delayed or off-line notes. More often gaps arise from my own lapses in writing (or at least being intelligible), though the automated records from Github in particular, as well as Flickr (image log), Mendeley (reading log), and Twitter and the like help make up for some of the gaps. The Integrated Notebook becomes the Knitted Notebook In creating my wordpress lab notebook, I put forward the idea of an “Integrated Lab Notebook”, a somewhat convoluted scheme in which I would describe my ideas and analyses in Wordpress posts, embed figures from Flickr, and link them to code on Github. Knitr simplified all that. I can now write code, analysis, figures, equations, citations, etc, into a single Rmarkdown format and track it’s evolution through git version control. The knitr markdown format goes smoothly on Github, the lab notebook, and even into generating pdf or word documents for publication, never seperating the code from the results. For details, see “writing reproducibly in the open with knitr.” Navigating the Open Notebook You can page through the notebook chronologically just like any paper notebook using the “Next” and “Previous” buttons on the sidebar. The notebook also leverages all of the standard features of a blog: the ability to search, browse the archives by date, browse by tag or browse by category. follow the RSS feed add and share comments in Disqus I use categories as the electronic equivalent of separate paper notebooks, dividing out my ecological research projects, evolutionary research topics, my teaching notebook, and a few others. As such, each entry is (usually) made into exactly one category. I use tags for more flexible topics, usually refecting particular projects or methods, and entries can have zero or multiple tags. It can be difficult to get the big picture of a project by merely flipping through entries. The chronological flow of a notebook is a poor fit to the very nonlinear nature of research. Reproducing particular results frequently requires additional information (also data and software) that are not part of the daily entries. Github repositories have been the perfect answer to these challenges. (The real notebook is Github) My Github repositories offer a kind of inverted version of the lab notebook, grouped by project (tag) rather than chronology. Each of my research projects is now is given it’s own public Github repository. I work primarily in R because it is widely used by ecologists and statisicians, and has a strong emphasis on reproducible research. The “R package” structure turns out to be brilliantly designed for research projects, which specifies particular files for essential metadata (title, description, authors, software dependencies, etc), data, documentation, and source code (see my workflow for details). Rather than have each analysis described in full in my notebook, they live as seperate knitr markdown files in the inst/examples directory of the R package, where their history can be browsed on Github, complete with their commit logs. Long or frequently used blocks of code are written into functions with proper documentation in the package source-code directory /R, keeping the analysis files cleaner and consistent. The issues tracker connected to each Github repository provides a rich TO DO list for the project. Progress on any issue often takes the form of subsequent commits of a particular analysis file, and that commit log can automatically be appended to the issue. The social lab notebook When scripting analyses or writing papers, pretty much everything can be captured on Github. I have recently added a short script to Jekyll which will pull the relevant commit logs into that day’s post automatically. Other activities fit less neatly into this mold (reading, math, notes from seminars and conferences), so these things get traditional notebook entries. I’m exploring automated integration for other activities, such as pulling my current reading from Mendeley or my recent discussions from Twitter into the notebook as well. For now, feed for each of these appear at the top of my notebook homepage, with links to the associated sites.
This emphasis on reproducibility matters to history too. It suggests I should keep detailed logs: where I got a manuscript image, how I interpreted marginalia, what uncertainties remain. That way future readers or researchers can trace my reasoning or redo steps themselves.
-
-
social-media-ethics-automation.github.io social-media-ethics-automation.github.io
-
20.7. Bibliography# [t1] Margaret Kohn and Kavita Reddy. Colonialism. In Edward N. Zalta and Uri Nodelman, editors, The Stanford Encyclopedia of Philosophy. Metaphysics Research Lab, Stanford University, spring 2023 edition, 2023. URL: https://plato.stanford.edu/archives/spr2023/entries/colonialism/ (visited on 2023-12-10). [t2] Hernán Cortés. November 2023. Page Version ID: 1186089050. URL: https://en.wikipedia.org/w/index.php?title=Hern%C3%A1n_Cort%C3%A9s&oldid=1186089050 (visited on 2023-12-10). [t3] Francisco Pizarro. December 2023. Page Version ID: 1188948507. URL: https://en.wikipedia.org/w/index.php?title=Francisco_Pizarro&oldid=1188948507 (visited on 2023-12-10). [t4] John Smith (explorer). December 2023. Page Version ID: 1189283105. URL: https://en.wikipedia.org/w/index.php?title=John_Smith_(explorer)&oldid=1189283105 (visited on 2023-12-10). [t5] Leopold II of Belgium. December 2023. Page Version ID: 1189115939. URL: https://en.wikipedia.org/w/index.php?title=Leopold_II_of_Belgium&oldid=1189115939 (visited on 2023-12-10). [t6] White savior. November 2023. Page Version ID: 1184795435. URL: https://en.wikipedia.org/w/index.php?title=White_savior&oldid=1184795435 (visited on 2023-12-10). [t7] Mighty Whitey. URL: https://tvtropes.org/pmwiki/pmwiki.php/Main/MightyWhitey (visited on 2023-12-10). [t8] White Man's Burden. URL: https://tvtropes.org/pmwiki/pmwiki.php/Main/WhiteMansBurden (visited on 2023-12-10). [t9] Ira Madison III. 'La La Land'’s White Jazz Narrative. MTV, December 2016. URL: https://www.mtv.com/news/5qr32e/la-la-lands-white-jazz-narrative (visited on 2023-12-10). [t10] Poster:The Last Samurai. February 2015. Page Version ID: 1025393048 This image is of a poster, and the copyright for it is most likely owned by either the publisher or the creator of the work depicted. URL: https://en.wikipedia.org/w/index.php?title=File:The_Last_Samurai.jpg&oldid=1025393048 (visited on 2023-12-10). [t11] The Last Samurai. December 2023. Page Version ID: 1188563405. URL: https://en.wikipedia.org/w/index.php?title=The_Last_Samurai&oldid=1188563405 (visited on 2023-12-10). [t12] Decolonization. December 2023. Page Version ID: 1189372296. URL: https://en.wikipedia.org/w/index.php?title=Decolonization&oldid=1189372296 (visited on 2023-12-10). [t13] Postcolonialism. November 2023. Page Version ID: 1186657050. URL: https://en.wikipedia.org/w/index.php?title=Postcolonialism&oldid=1186657050 (visited on 2023-12-10). [t14] Liberation movement. October 2023. Page Version ID: 1180933418. URL: https://en.wikipedia.org/w/index.php?title=Liberation_movement&oldid=1180933418 (visited on 2023-12-10). [t15] Land Back. December 2023. Page Version ID: 1188237630. URL: https://en.wikipedia.org/w/index.php?title=Land_Back&oldid=1188237630 (visited on 2023-12-10). [t16] Mahatma Gandhi. December 2023. Page Version ID: 1189603306. URL: https://en.wikipedia.org/w/index.php?title=Mahatma_Gandhi&oldid=1189603306 (visited on 2023-12-10). [t17] Toussaint Louverture. November 2023. Page Version ID: 1187587809. URL: https://en.wikipedia.org/w/index.php?title=Toussaint_Louverture&oldid=1187587809 (visited on 2023-12-10). [t18] Patrice Lumumba. December 2023. Page Version ID: 1189622266. URL: https://en.wikipedia.org/w/index.php?title=Patrice_Lumumba&oldid=1189622266 (visited on 2023-12-10). [t19] Susan B. Anthony. December 2023. Page Version ID: 1188464282. URL: https://en.wikipedia.org/w/index.php?title=Susan_B._Anthony&oldid=1188464282 (visited on 2023-12-10). [t20] Martin Luther King Jr. December 2023. Page Version ID: 1188881438. URL: https://en.wikipedia.org/w/index.php?title=Martin_Luther_King_Jr.&oldid=1188881438 (visited on 2023-12-10). [t21] Nelson Mandela. December 2023. Page Version ID: 1188461215. URL: https://en.wikipedia.org/w/index.php?title=Nelson_Mandela&oldid=1188461215 (visited on 2023-12-10). [t22] Gayatri Chakravorty Spivak. December 2023. Page Version ID: 1189060723. URL: https://en.wikipedia.org/w/index.php?title=Gayatri_Chakravorty_Spivak&oldid=1189060723 (visited on 2023-12-10). [t23] Edward Said. November 2023. Page Version ID: 1187438394. URL: https://en.wikipedia.org/w/index.php?title=Edward_Said&oldid=1187438394 (visited on 2023-12-10). [t24] One Laptop per Child. November 2023. Page Version ID: 1187517049. URL: https://en.wikipedia.org/w/index.php?title=One_Laptop_per_Child&oldid=1187517049 (visited on 2023-12-10). [t25] Adi Robertson. OLPC’s \$100 laptop was going to change the world — then it all went wrong. The Verge, April 2018. URL: https://www.theverge.com/2018/4/16/17233946/olpcs-100-laptop-education-where-is-it-now (visited on 2023-12-10). [t26] Non-English-based programming languages. November 2023. Page Version ID: 1185172571. URL: https://en.wikipedia.org/w/index.php?title=Non-English-based_programming_languages&oldid=1185172571 (visited on 2023-12-10). [t27] Philip J. Guo. Non-Native English Speakers Learning Computer Programming: Barriers, Desires, and Design Opportunities. In Proceedings of the 2018 CHI Conference on Human Factors in Computing Systems, CHI '18, 1–14. New York, NY, USA, April 2018. Association for Computing Machinery. URL: https://doi.org/10.1145/3173574.3173970 (visited on 2023-12-12), doi:10.1145/3173574.3173970. [t28] Yuri Takhteyev. Coding Places: Software Practice in a South American City. September 2012. URL: https://mitpress.mit.edu/9780262018074/coding-places/ (visited on 2023-12-10), doi:10.7551/mitpress/9109.001.0001. [t29] David Robinson. A Tale of Two Industries: How Programming Languages Differ Between Wealthy and Developing Countries - Stack Overflow. August 2017. URL: https://stackoverflow.blog/2017/08/29/tale-two-industries-programming-languages-differ-wealthy-developing-countries/ (visited on 2023-12-10). [t30] Lua (programming language). December 2023. Page Version ID: 1189590273. URL: https://en.wikipedia.org/w/index.php?title=Lua_(programming_language)&oldid=1189590273 (visited on 2023-12-10). [t31] Lev Grossman. Exclusive: Inside Facebook’s Plan to Wire the World. Time, December 2014. URL: https://time.com/facebook-world-plan/ (visited on 2023-12-10). [t32] The Hitchhiker's Guide to the Galaxy (novel). November 2023. Page Version ID: 1184131911. URL: https://en.wikipedia.org/w/index.php?title=The_Hitchhiker%27s_Guide_to_the_Galaxy_(novel)&oldid=1184131911 (visited on 2023-12-10). [t33] Dan Milmo. Rohingya sue Facebook for £150bn over Myanmar genocide. The Guardian, December 2021. URL: https://www.theguardian.com/technology/2021/dec/06/rohingya-sue-facebook-myanmar-genocide-us-uk-legal-action-social-media-violence (visited on 2023-12-10). [t34] Craig Silverman, Craig Timberg, Jeff Kao, and Jeremy Merrill. Facebook Hosted Surge of Misinformation and Insurrection Threats in Months Leading Up to Jan. 6 Attack, Records Show. ProPublica, January 2022. URL: https://www.propublica.org/article/facebook-hosted-surge-of-misinformation-and-insurrection-threats-in-months-leading-up-to-jan-6-attack-records-show (visited on 2023-12-10). [t35] Mark Zuckerberg. Bringing the world closer together. March 2021. URL: https://www.facebook.com/notes/393134628500376/ (visited on 2023-12-10). [t36] Meta - Resources. 2022. URL: https://investor.fb.com/resources/default.aspx (visited on 2023-12-10). [t37] Olivia Solon. 'It's digital colonialism': how Facebook's free internet service has failed its users. The Guardian, July 2017. URL: https://www.theguardian.com/technology/2017/jul/27/facebook-free-basics-developing-markets (visited on 2023-12-10). [t38] Josh Constine and Kim-Mai Cutler. Why Facebook Dropped \$19B On WhatsApp: Reach Into Europe, Emerging Markets. TechCrunch, February 2014. URL: https://techcrunch.com/2014/02/19/facebook-whatsapp/ (visited on 2023-12-10). { requestKernel: true, binderOptions: { repo: "binder-examples/jupyter-stacks-datascience", ref: "master", }, codeMirrorConfig: { theme: "abcdef", mode: "python" }, kernelOptions: { name: "python3", path: "./ch20_colonialism" }, predefinedOutput: true } kernelName = 'python3'
One source that stood out to me was the StackOverflow study (t29) about how programming languages differ between wealthy and developing countries. The most interesting detail I learned from that article is that Python and R—two languages I always hear people hype up—are barely used in poorer countries. Meanwhile, older languages like PHP and Android development stay extremely common there. The study explains that it’s not because developers in those countries “prefer” outdated tech, but because the global tech industry is shaped around Silicon Valley’s needs. That really clicked for me. It shows how something as simple as a programming language choice is actually influenced by economics and access, not just technical preference. It made me rethink the whole idea that tech is some neutral, equal-opportunity field.
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
A major point all three reviewers raise is that the ‘human-AI collaboration’ in our experiment may not be true collaboration (as the AI does not classify images per se), but that it is only implied. The reviewers pointed out that whether participants were genuinely engaged in our experimental task is currently not sufficiently addressed. We plan to address this issue in the revised manuscript by including results from a brief interview we conducted after the experiment with each participant, which asked about the participant’s experience and decision-making processes while performing the task. Additionally, we also measured the participants’ propensity to trust in AI via a questionnaire before and after the experiment. The questionnaire and interview results will allow us to more accurately describe the involvement of our participants in the task. Additionally, we will conduct additional analyses of the behavioural data (e.g., response times) to show that participants genuinely completed the experimental task. Finally, we will work to sharpen our language and conclusions in the revised manuscript, following the reviewers’ recommendations.
Reviewer #1:
Summary:
In the study by Roeder and colleagues, the authors aim to identify the psychophysiological markers of trust during the evaluation of matching or mismatching AI decision-making. Specifically, they aim to characterize through brain activity how the decision made by an AI can be monitored throughout time in a two-step decision-making task. The objective of this study is to unfold, through continuous brain activity recording, the general information processing sequence while interacting with an artificial agent, and how internal as well as external information interact and modify this processing. Additionally, the authors provide a subset of factors affecting this information processing for both decisions.
Strengths:
The study addresses a wide and important topic of the value attributed to AI decisions and their impact on our own confidence in decision-making. It especially questions some of the factors modulating the dynamical adaptation of trust in AI decisions. Factors such as perceived reliability, type of image, mismatch, or participants' bias toward one response or the other are very relevant to the question in human-AI interactions.
Interestingly, the authors also question the processing of more ambiguous stimuli, with no real ground truth. This gets closer to everyday life situations where people have to make decisions in uncertain environments. Having a better understanding of how those decisions are made is very relevant in many domains.
Also, the method for processing behavioural and especially EEG data is overall very robust and is what is currently recommended for statistical analyses for group studies. Additionally, authors provide complete figures with all robustness evaluation information. The results and statistics are very detailed. This promotes confidence, but also replicability of results.
An additional interesting method aspect is that it is addressing a large window of analysis and the interaction between three timeframes (evidence accumulation pre-decision, decision-making, post-AI decision processing) within the same trials. This type of analysis is quite innovative in the sense that it is not yet a standard in complex experimental designs. It moves forward from classical short-time windows and baseline ERP analysis.
We appreciate the constructive appraisal of our work.
Weaknesses:
R1.1. This manuscript raises several conceptual and theoretical considerations that are not necessarily answered by the methods (especially the task) used. Even though the authors propose to assess trust dynamics and violations in cooperative human-AI teaming decision-making, I don't believe their task resolves such a question. Indeed, there is no direct link between the human decision and the AI decision. They do not cooperate per se, and the AI decision doesn't seem, from what I understood to have an impact on the participants' decision making. The authors make several assumptions regarding trust, feedback, response expectation, and "classification" (i.e., match vs. mismatch) which seem far stretched when considering the scientific literature on these topics.
This issue is raised by the other reviewers as well. The reviewer is correct in that the AI does not classify images but that the AI response is dependent on the participants’ choice (agree in 75% of trials, disagree in 25% of the trials). Importantly, though, participants were briefed before and during the experiment that the AI is doing its own independent image classification and that human input is needed to assess how well the AI image classification works. That is, participants were led to believe in a genuine, independent AI image classifier on this experiment.
Moreover, the images we presented in the experiment were taken from previous work by Nightingale & Farid (2022). This image dataset includes ‘fake’ (AI generated) images that are indistinguishable from real images.
What matters most for our work is that the participants were truly engaging in the experimental task; that is, they were genuinely judging face images, and they were genuinely evaluating the AI feedback. There is strong indication that this was indeed the case. We conducted and recorded brief interviews after the experiment, asking our participants about their experience and decision-making processes. The questions are as follows:
(1) How did you make the judgements about the images?
(2) How confident were you about your judgement?
(3) What did you feel when you saw the AI response?
(4) Did that change during the trials?
(5) Who do you think it was correct?
(6) Did you feel surprised at any of the AI responses?
(7) How did you judge what to put for the reliability sliders?
In our revised manuscript we will conduct additional analyses to provide detail on participants’ engagement in the task; both in the judging of the AI faces, as well as in considering the AI feedback. In addition, we will investigate the EEG signal and response time to check for effects that carry over between trials. We will also frame our findings more carefully taking scientific literature into account.
Nightingale SJ, and Farid H. "AI-synthesized faces are indistinguishable from real faces and more trustworthy." Proceedings of the National Academy of Sciences 119.8 (2022): e2120481119.
R1.2. Unlike what is done for the data processing, the authors have not managed to take the big picture of the theoretical implications of their results. A big part of this study's interpretation aims to have their results fit into the theoretical box of the neural markers of performance monitoring.
We indeed used primarily the theoretical box of performance monitoring and predictive coding, since the make-up of our task is similar to a more classical EEG oddball paradigm. In our revised manuscript, we will re-frame and address the link of our findings with the theoretical framework of evidence accumulation and decision confidence.
R1.3. Overall, the analysis method was very robust and well-managed, but the experimental task they have set up does not allow to support their claim. Here, they seem to be assessing the impact of a mismatch between two independent decisions.
Although the human and AI decisions are independent in the current experiment, the EEG results still shed light on the participant’s neural processes, as long as the participant considers the AI’s decision and believes it to be genuine. An experiment in which both decisions carry effective consequences for the task and the human-AI cooperation would be an interesting follow-up study.
Nevertheless, this type of work is very important to various communities. First, it addresses topical concerns associated with the introduction of AI in our daily life and decisions, but it also addresses methodological difficulties that the EEG community has been having to move slowly away from the static event-based short-timeframe analyses onto a more dynamic evaluation of the unfolding of cognitive processes and their interactions. The topic of trust toward AI in cooperative decision making has also been raised by many communities, and understanding the dynamics of trust, as well as the factors modulating it, is of concern to many high-risk environments, or even everyday life contexts. Policy makers are especially interested in this kind of research output.
Reviewer #2:
Summary:
The authors investigated how "AI-agent" feedback is perceived in an ambiguous classification task, and categorised the neural responses to this. They asked participants to classify real or fake faces, and presented an AI-agent's feedback afterwards, where the AI-feedback disagreed with the participants' response on a random 25% of trials (called mismatches). Pre-response ERP was sensitive to participants' classification as real or fake, while ERPs after the AI-feedback were sensitive to AI-mismatches, with stronger N2 and P3a&b components. There was an interaction of these effects, with mismatches after a "Fake" response affecting the N2 and those after "Real" responses affecting P3a&b. The ERPs were also sensitive to the participants' response biases, and their subjective ratings of the AI agent's reliability.
Strengths:
The researchers address an interesting question, and extend the AI-feedback paradigm to ambiguous tasks without veridical feedback, which is closer to many real-world tasks. The in-depth analysis of ERPs provides a detailed categorisation of several ERPs, as well as whole-brain responses, to AI-feedback, and how this interacts with internal beliefs, response biases, and trust in the AI-agent.
We thank the reviewer for their time in reading and reviewing our manuscript.
Weaknesses:
R2.1. There is little discussion of how the poor performance (close to 50% chance) may have affected performance on the task, such as by leading to entirely random guessing or overreliance on response biases. This can change how error-monitoring signals presented, as they are affected by participants' accuracy, as well as affecting how the AI feedback is perceived.
The images were chosen from a previous study (Nightingale & Farid, 2022, PNAS) that looked specifically at performance accuracy and also found levels around 50%. Hence, ‘fake’ and ‘real’ images are indistinguishable in this image dataset. Our findings agree with the original study.
Judging based on the brief interviews after the experiment (see answer to R.1.1.), all participants were actively and genuinely engaged in the task, hence, it is unlikely that they pressed buttons at random. As mentioned above, we will include a formal analysis of the interviews in the revised manuscript.
The response bias might indeed play a role in how participants responded, and this might be related to their initial propensity to trust in AI. We have questionnaire data available that might shed light on this issue: before and after the experiment, all participants answered the following questions with a 5-point Likert scale ranging from ‘Not True’ to ‘Completely True’:
(1) Generally, I trust AI.
(2) AI helps me solve many problems.
(3) I think it's a good idea to rely on AI for help.
(4) I don't trust the information I get from AI.
(5) AI is reliable.
(6) I rely on AI.
The propensity to trust questionnaire is adapted from Jessup SA, Schneider T R, Alarcon GM, Ryan TJ, & Capiola A. (2019). The measurement of the propensity to trust automation. International Conference on Human-Computer Interaction.
Our initial analyses did not find a strong link between the initial (before the experiment) responses to these questions, and how images were rated during the experiment. We will re-visit this analysis and add the results to the revised manuscript.
Regarding how error-monitoring (or the equivalent thereof in our experiment) is perceived, we will analyse interview questions 3 (“What did you feel when you saw the AI response”) and 6 (“Did you feel surprised at any of the AI responses”) and add results to the revised manuscript.
The task design and performance make it hard to assess how much it was truly measuring "trust" in an AI agent's feedback. The AI-feedback is yoked to the participants' performance, agreeing on 75% of trials and disagreeing on 25% (randomly), which is an important difference from the framing provided of human-AI partnerships, where AI-agents usually act independently from the humans and thus disagreements offer information about the human's own performance. In this task, disagreements are uninformative, and coupled with the at-chance performance on an ambiguous task, it is not clear how participants should be interpreting disagreements, and whether they treat it like receiving feedback about the accuracy of their choices, or whether they realise it is uninformative. Much greater discussion and justification are needed about the behaviour in the task, how participants did/should treat the feedback, and how these affect the trust/reliability ratings, as these are all central to the claims of the paper.
In our experiment, the AI disagreements are indeed uninformative for the purpose of making a correct judgment (that is, correctly classifying images as real or fake). However, given that the AI-generated faces are so realistic and indistinguishable from the real faces, the correctness of the judgement is not the main experimental factor in this study. We argue that, provided participants were genuinely engaged in the task, their judgment accuracy is less important than their internal experience when the goal is to examine processes occurring within the participants themselves. We briefed our participants as follows before the experiment:
“Technology can now create hyper-realistic images of people that do not exist. We are interested in your view on how well our AI system performs at identifying whether images of people’s faces are real or fake (computer-generated). Human input is needed to determine when a face looks real or fake. You will be asked to rate images as real or fake. The AI system will also independently rate the images. You will rate how reliable the AI is several times throughout the experiment.”
We plan to more fully expand the behavioural aspect and our participants’ experience in the revised manuscript by reporting the brief post-experiment interview (R.1.1.), the propensity to trust questionnaire (R.2.1.), and additional analyses of the response times.
There are a lot of EEG results presented here, including whole-brain and window-free analyses, so greater clarity on which results were a priori hypothesised should be given, along with details on how electrodes were selected for ERPs and follow-up tests.
We chose the electrodes mainly to be consistent across findings, and opted to use central electrodes (Pz and Fz), as long as the electrode was part of the electrodes within the reported cluster. We can in our revised manuscript also report on the electrodes with the maximal statistic, as part of a more complete and descriptive overview. We will also report on where we expected to see ERP components within the paper. In short, we did expect something like a P3, and we did also expect to see something before the response what we call the CPP. The rest of the work was more exploratory, with a more careful expectation that bias would be connected to the CPP, and the reliability ratings more to the P3; however, we find the opposite results. We will include this in our revised work as well.
We selected the electrodes primarily to maintain consistency across our findings and figures, and focused on central electrodes (Pz and Fz), provided they fell within the reported cluster. In the revised manuscript, we will also report the electrodes showing the maximal statistical effects to give a more complete and descriptive overview. Additionally, we will report where we expected specific ERP components to appear. In brief, we expected to see a P3 component post AI feedback, and a pre-response signal corresponding to the CPP. Beyond these expectations, the remaining analyses were more exploratory. Although we tentatively expected bias to relate to the CPP and reliability ratings to the P3, our results showed the opposite pattern. We will clarify this in the revised version of the manuscript.
Reviewer #3:
The current paper investigates neural correlates of trust development in human-AI interaction, looking at EEG signatures locked to the moment that AI advice is presented. The key finding is that both human-response-locked EEG signatures (the CPP) and post-AI-advice signatures (N2, P3) are modulated by trust ratings. The study is interesting, however, it does have some clear and sometimes problematic weaknesses:
(1) The authors did not include "AI-advice". Instead, a manikin turned green or blue, which was framed as AI advice. It is unclear whether participants viewed this as actual AI advice.
This point has been raised by the other reviewers as well, and we refer to the answers under R1.1., and under R2.1. We will address this concern by analysing the post-experiment interviews. In particular, questions 3 (“What did you feel when you saw the AI response”), 4 (“Did that change during the trials?”) and 6 (“Did you feel surprised at any of the AI responses”) will give critical insight. As stated above, our general impression from conducting the interviews is that all participants considered the robot icon as decision from an independent AI agent.
(2) The authors did not include a "non-AI" control condition in their experiment, such that we cannot know how specific all of these effects are to AI, or just generic uncertain feedback processing.
In the conceptualization phase of this study, we indeed considered different control conditions for our experiment to contrast different kinds of feedback. However, previous EEG studies on performance monitoring ERPs have reported similar results for human and machine supervision (Somon et al., 2019; de Visser et al., 2018). We therefore decided to focus on one aspect (the judgement of observation of an AI classification), also to prevent the experiment from taking too long and risking that participants would lose concentration and motivation to complete the experiment. Comparing AI vs non-AI feedback, is still interesting and would be a valuable follow-up study.
Somon B, et al. "Human or not human? Performance monitoring ERPs during human agent and machine supervision." NeuroImage 186 (2019): 266-277.
De Visser EJ, et al. "Learning from the slips of others: Neural correlates of trust in automated agents." Frontiers in human neuroscience 12 (2018): 309.
(3) Participants perform the task at chance level. This makes it unclear to what extent they even tried to perform the task or just randomly pressed buttons. These situations likely differ substantially from a real-life scenario where humans perform an actual task (which is not impossible) and receive actual AI advice.
This concern was also raised by the other two reviewers. As already stated in our responses above, we will add results from the post-experiment interviews with the participants, the propensity to trust questionnaire, and additional behavioural analyses in our revised manuscript.
Reviewer 1 (R1.3) also brought up the situation where decisions by the participant and the AI have a more direct link which carries consequences. This will be valuable follow-up research. In the revised manuscript, we will more carefully frame our approach.
(4) Many of the conclusions in the paper are overstated or very generic.
In the revised manuscript, we will re-phrase our discussion and conclusions to address the points raised in the reviewer’s recommendations to authors.
-
-
www.biorxiv.org www.biorxiv.org
-
Reviewer #1 (Public review):
Summary:
This manuscript provides a comprehensive systematic analysis of envelope-containing Ty3/gypsy retrotransposons (errantiviruses) across metazoan genomes, including both invertebrates and ancient animal lineages. Using iterative tBLASTn mining of over 1,900 genomes, the authors catalog 1,512 intact retrotransposons with uninterrupted gag, pol, and env open reading frames. They show that these elements are widespread-present in most metazoan phyla, including cnidarians, ctenophores, and tunicates-with active proliferation indicated by their multicopy status. Phylogenetic analyses distinguish "ancient" and "insect" errantivirus clades, while structural characterization (including AlphaFold2 modeling) reveals two major env types: paramyxovirus F-like and herpesvirus gB-like proteins. Although bot envelope types were identified in previous analyses two decades ago, the evolutionary provenance of these envelope genes was almost rudimentary and anecdotal (I can say this because I authored one of these studies). The results in the present study support an ancient origin for env acquisition in metazoan Ty3/gypsy elements, with subsequent vertical inheritance and limited recombination between env and pol domains. The paper also proposes an expanded definition of 'errantivirus' for env-carrying Ty3/gypsy elements outside Drosophila.
Strengths:
(1) Comprehensive Genomic Survey:<br /> The breadth of the genome search across non-model metazoan phyla yields an impressive dataset covering evolutionary breadth, with clear documentation of search iterations and validation criteria for intact elements.
(2) Robust Phylogenetic Inference:<br /> The use of maximum likelihood trees on both pol and env domains, with thorough congruence analysis, convincingly separates ancient from lineage-specific elements and demonstrates co-evolution of env and pol within clades.
(3) Structural Insights:<br /> AlphaFold2-based predictions provide high-confidence structural evidence that both env types have retained fusion-competent architectures, supporting the hypothesis of preserved functional potential.
(4) Novelty and Scope:<br /> The study challenges previous assumptions of insect-centric or recent env acquisition and makes a compelling case for a Pre-Cambrian origin, significantly advancing our understanding of animal retroelement diversity and evolution. THIS IS A MAJOR ADVANCE.
(5) Data Transparency:<br /> I appreciate that all data, code, and predicted structures are made openly available, facilitating reproducibility and future comparative analyses.
Major Weaknesses
(1) Functional Evidence Gaps:<br /> The work rests largely on sequence and structure prediction. No direct expression or experimental validation of envelope gene function or infectivity outside Drosophila is attempted, which would be valuable to corroborate the inferred roles of these glycoproteins in non-insect lineages. At least for some of these species, there are RNA-seq datasets that could be leveraged.
(2) Horizontal Transfer vs. Loss Hypotheses:<br /> The discussion argues primarily for vertical inheritance, but the somewhat sporadic phylogenetic distributions and long-branch effects suggest that loss and possibly rare horizontal events may contribute more than acknowledged. Explicit quantitative tests for horizontal transfer, or reconciliation analyses, would strengthen this conclusion. It's also worth pointing out that, unlike retrotransposons that can be found in genomes, any potential related viral envelopes must, by definition, have a spottier distribution due to sampling. I don't think this challenges any of the conclusions, but it must be acknowledged as something that could affect the strength of this conclusion
(3) Limited Taxon Sampling for Certain Phyla:<br /> Despite the impressive breadth, some ancient lineages (e.g., Porifera, Echinodermata) are negative, but the manuscript does not fully explore whether this reflects real biological absence, assembly quality, or insufficient sampling. A more systematic treatment of negative findings would clarify claims of ubiquity. However, I also believe this falls beyond the scope of this study.
(4) Mechanistic Ambiguity:<br /> The proposed model that env-containing elements exploit ovarian somatic niches is plausible but extrapolated from Drosophila data; for most taxa, actual tissue specificity, lifecycle, or host interaction mechanisms remain speculative and, to me, a bit unreasonable.
Minor Weaknesses:
(1) Terminology and Nomenclature:<br /> The paper introduces and then generalizes the term "errantivirus" to non-insect elements. While this is logical, it may confuse readers familiar with the established, Drosophila-centric definition if not more explicitly clarified throughout. I also worry about changes being made without any input from the ICTV nomenclature committee, which just went through a thorough reclassification. Nevertheless, change is expected, and calling them all errantiviruses is entirely reasonable.
(2) Figures and Supplementary Data Navigation:<br /> Some key phylogenies and domain alignments are found only in supplementary figures, occasionally hindering readability for non-expert audiences. Selected main-text inclusion of representative trees would benefit accessibility.
(3) ORF Integrity Thresholds:<br /> The cutoff choices for defining "intact" elements (e.g., numbers/placement of stop codons, length ranges) are reasonable but only lightly justified. More rationale or sensitivity analysis would improve confidence in the inclusion criteria. For example, how did changing these criteria change the number of intact elements?
(4) Minor Typos/Formatting:<br /> The paper contains sporadic typographical errors and formatting glitches (e.g., misaligned figure labels, unrendered symbols) that should be addressed.
-
- Nov 2025
-
www.mediaed.org www.mediaed.org
-
when we are immersed in something, surrounded by it the waywe are by images from the media, we may come to accept them as just part ofthe real and natural world.
This line makes sense to me because it explains how easy it is to stop questioning the media we see everyday. When something is constantly shown, like stereotypes in movies or the way certain groups are shown. It starts to feel normal even if it's totally inaccurate. This makes Hall's point, that we have to step back and actually think about what we're being shown instead of just absorbing it without realizing, clear.
-
-
autre.love autre.love
-
My philosophy of authenticity is that it doesn’t exist in the way people wish it did. I don’t believe it’s possible to perform in a way that’s authentic. People will say, I just post for myself, which is a lie. They say that because they feel it’s morally better to be that way, and I really disagree with that. It’s okay to feel like you’re performing and even want to perform a bit. That’s not evil. It’s a condition of living. I’ve adopted a [Erving] Goffman-esque philosophy of performance online. Everything is a performance. Goffman was writing before the internet, so he is talking about socializing in general, which I also think is true. It’s been kind of freeing for me to subscribe to this notion that authenticity does not exist.
not possible to perform in a way that's authentic vs. authenticity not existing – two slightly different ideas
moreover, though: mastodon getting stuck, posting anyway
-
-
social-media-ethics-automation.github.io social-media-ethics-automation.github.io
-
Merriam-Webster. Definition of SOCIALISM. December 2023. URL: https://www.merriam-webster.com/dictionary/socialism (visited on 2023-12-10).
When I looked up socialism in Merriam Webster, I noticed how clearly it explained the idea of shared ownership and public control of resources. It helped me understand that socialism isn’t just about government involvement, it’s about distributing power and resources more equally. This definition made me think about how different the system feels compared to capitalism, where individual ownership and profit are the main focus.
-
-
hypothes.is hypothes.is
-
1— “Debate has raged”
Some headline news from the budget: Labour is finally, after an 18-month internal battle, scrapping the two-child benefit cap. How did they get here? Ailbhe is here, as always, with the inside track. Finn
2—“Mortal danger”
Is it all over in Ukraine? The country cannot fight a war for another year, that much is clear. Europe is facing a lonely future, without its American guarantor and with an expansionist, unchecked Russia. Andrew Marr assesses the grave situation. Finn
3—“How did this happen?”
Will Dunn makes an unappetising expedition for the sketch this week. There is “a hulking glacier of crap 500 feet long in the heart of the Oxfordshire countryside.” Criminals used it as an illegal rubbish tip. Will holds his nose and follows Ed Davey once more unto the heap. George
4—“Her rally or his…”
It’s Your Party conference weekend, and it’s going to be massive. Some predict a barney, some a bust-up. We’ve got two pieces for the meantime. First, Megan Kenyon sat down with Jeremy Corbyn to discuss his apology to Your Party members, his breakfast meeting with Zack Polanski and his ambitions for the leadership. Watch here, and read here.
And then we have a weekend essay from the left-wing veteran, Andrew Murray. He has some advice for the Your Party high-ups, most saliently to “to stop doing stupid stuff”. Nicholas
5—“Who was Salman Rushdie?”
This is a major one. When one colleague asked Tanjil how he felt to be writing about Sir Salman Rushdie, he said, “Well, I have been reading him since I was a boy.” And Tanjil’s boyhood is foreground and background in this essay-cum-meditation-cum-memoir. Not a dry eye in the house. Nicholas
To enjoy our latest analysis of politics, news and events, in addition to world-class literary and cultural reviews, click here to subscribe to the New Statesman. You'll enjoy all of the New Statesman's online content, ad-free podcasts and invitations to NS events.
75% off
6—”Here’s the trick”
It takes a village (or un village?). While Will Dunn was inspecting the giant trash heap I was thoroughly investigating this year’s Beaujolais nouveau. Come along for a glass of summer in the bleak mid winter: the unassuming Gamay grape can teach us more than you might think about life. Trust me, or read me, to find out what. Finn
7—“Hymns of isolation”
I’ve always thought of Radiohead as headphone music: that falsetto over those arrangements, it’s something intense and private, not for 20,000 people standing in a field. But, in this wonderful review of the band live, George has won me round to the alternative. Nicholas
8—”Just-so satisfaction” William Nicholson and the pleasure in the paint No one can really agree on how significant William Nicholson’s contribution to 20th century painting was. Probably thanks to all those plodding still lifes. Michael Prodger jumps in to tell me to stop being such a hater – there is real pleasure in the close reading, he says. Convinced? Finn
9—”Like the Stasi in East Berlin”
Ethan Croft scopes out a faction with traction in the Labour party. Blue Labour involves a “bricolage of calls for reindustrialisation and lower migration, inspired by Catholic social teaching”. Others write it off as a load of Tories. Its influence has gone up, then down, then up, and so on. Right now they’re riding high. Ethan never fails to provide your quotient of gossip and Labour infighting. George
Elsewhere Naomi Klein: surrealism against fascism (from the brilliant new mag, Equator)
Why would China want to trade with us?
Guardian investigates the Free Birth Society
New Yorker: Airport lounge wars
Atlantic: Stranger Things comes to an exhausting end
Ryan Lizza/Olivia Nuzzi latest
Gamma the tortoise dies in her prime, at 141 :(
Recipe of the week: Nigel Slater’s pear and chocolate crumble (a crowd pleaser)
And with that…
Something smells fishy! And snail-y. And wine-y. I am talking, of course, about the recent spate of luxury grocery theft. Some thieves have stolen €90,000 worth of snails, intended for the restaurant trade. The producer (funny word for that job, I thought) said he was shocked when he learnt of the disappearance of 450kg of snails from his farm in Bouzy, in – get this – the Champagne region of France. The Times described the theft as “yet another blow to a struggling sector”.
Meanwhile, closer to home in Chelsea, a woman has been caught on CCTV making off with a box of langoustines, stolen from the doorstep of the Michelin-starred restaurant Elystan Street. That’s about £200 worth of big prawns. And in Virginia, a couple posed as wealthy collectors in order to secure private tours of restaurant wine cellars. While one distracted the sommelier, the other swiped. In their haul? A rare 2020 Romanée-Conti, worth $24,000.
I can’t help but think about the Louvre jewel heist in October: a crime of extraordinary effort. To pull it off, you do not just need to outsmart Louvre security, you then have to work out how to sell the things. And as Michael explains, flogging stolen jewels without alerting the authorities is a hard task. Snail theft is starting to sound appealing: no need for a cross-border pan-European crime network or experts in recutting precious stones; just a hot oven, some salted butter, chopped parsley and a splash of dry white, and you have already succeeded.
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public review):
Summary:
From a forward genetic mosaic mutant screen using EMS, the authors identify mutations in glucosylceramide synthase (GlcT), a rate-limiting enzyme for glycosphingolipid (GSL) production, that result in EE tumors. Multiple genetic experiments strongly support the model that the mutant phenotype caused by GlcT loss is due to by failure of conversion of ceramide into glucosylceramide. Further genetic evidence suggests that Notch signaling is comprised in the ISC lineage and may affect the endocytosis of Delta. Loss of GlcT does not affect wing development or oogenesis, suggesting tissue-specific roles for GlcT. Finally, an increase in goblet cells in UGCG knockout mice, not previously reported, suggests a conserved role for GlcT in Notch signaling in intestinal cell lineage specification.
Strengths:
Overall, this is a well-written paper with multiple well-designed and executed genetic experiments that support a role for GlcT in Notch signaling in the fly and mammalian intestine. I do, however, have a few comments below.
Weaknesses:
(1) The authors bring up the intriguing idea that GlcT could be a way to link diet to cell fate choice. Unfortunately, there are no experiments to test this hypothesis.
We indeed attempted to establish an assay to investigate the impact of various diets (such as high-fat, high-sugar, or high-protein diets) on the fate choice of ISCs. Subsequently, we intended to examine the potential involvement of GlcT in this process. However, we observed that the number or percentage of EEs varies significantly among individuals, even among flies with identical phenotypes subjected to the same nutritional regimen. We suspect that the proliferative status of ISCs and the turnover rate of EEs may significantly influence the number of EEs present in the intestinal epithelium, complicating the interpretation of our results. Consequently, we are unable to conduct this experiment at this time. The hypothesis suggesting that GlcT may link diet to cell fate choice remains an avenue for future experimental exploration.
(2) Why do the authors think that UCCG knockout results in goblet cell excess and not in the other secretory cell types?
This is indeed an interesting point. In the mouse intestine, it is well-documented that the knockout of Notch receptors or Delta-like ligands results in a classic phenotype characterized by goblet cell hyperplasia, with little impact on the other secretory cell types. This finding aligns very well with our experimental results, as we noted that the numbers of Paneth cells and enteroendocrine cells appear to be largely normal in UGCG knockout mice. By contrast, increases in other secretory cell types are typically observed under conditions of pharmacological inhibition of the Notch pathway.
(3) The authors should cite other EMS mutagenesis screens done in the fly intestine.
To our knowledge, the EMS screen on 2L chromosome conducted in Allison Bardin’s lab is the only one prior to this work, which leads to two publications (Perdigoto et al., 2011; Gervais, et al., 2019). We have now included citations for both papers in the revised manuscript.
(4) The absence of a phenotype using NRE-Gal4 is not convincing. This is because the delay in its expression could be after the requirement for the affected gene in the process being studied. In other words, sufficient knockdown of GlcT by RNA would not be achieved until after the relevant signaling between the EB and the ISC occurred. Dl-Gal4 is problematic as an ISC driver because Dl is expressed in the EEP.
This is an excellent point, and we agree that the lack of an observable phenotype using NRE-Gal4 could be due to delayed expression, which may result in missing the critical window required for effective GlcT knockdown. Consequently, we cannot rule out the possibility that GlcT also plays a role in early EBs or EEPs. We have revised the manuscript to soften this conclusion and to include this alternative explanation for the experiment.
(5) The difference in Rab5 between control and GlcT-IR was not that significant. Furthermore, any changes could be secondary to increases in proliferation.
We agree that it is possible that the observed increase in proliferation could influence the number of Rab5+ endosomes, and we will temper our conclusions on this aspect accordingly. However, it is important to note that, although the difference in Rab5+ endosomes between the control and GlcT-IR conditions appeared mild, it was statistically significant and reproducible. In our revised experiments, we have not only added statistical data and immunofluorescence images for Rab11 but also unified the approaches used for detecting Rab-associated proteins (in the previous figures, Rab5 was shown using U-Rab5-GFP, whereas Rab7 was detected by direct antibody staining). Based on this unified strategy, we optimized the quantification of Dl-GFP colocalization with early, late, and recycling endosomes, and the results are consistent with our previous observations (see the updated Fig. 5).
Reviewer #2 (Public review):
Summary:
This study genetically identifies two key enzymes involved in the biosynthesis of glycosphingolipids, GlcT and Egh, which act as tumor suppressors in the adult fly gut. Detailed genetic analysis indicates that a deficiency in Mactosyl-ceramide (Mac-Cer) is causing tumor formation. Analysis of a Notch transcriptional reporter further indicates that the lack of Mac-Ser is associated with reduced Notch activity in the gut, but not in other tissues.
Addressing how a change in the lipid composition of the membranes might lead to defective Notch receptor activation, the authors studied the endocytic trafficking of Delta and claimed that internalized Delta appeared to accumulate faster into endosomes in the absence of Mac-Cer. Further analysis of Delta steady-state accumulation in fixed samples suggested a delay in the endosomal trafficking of Delta from Rab5+ to Rab7+ endosomes, which was interpreted to suggest that the inefficient, or delayed, recycling of Delta might cause a loss in Notch receptor activation.
Finally, the histological analysis of mouse guts following the conditional knock-out of the GlcT gene suggested that Mac-Cer might also be important for proper Notch signaling activity in that context.
Strengths:
The genetic analysis is of high quality. The finding that a Mac-Cer deficiency results in reduced Notch activity in the fly gut is important and fully convincing.
The mouse data, although preliminary, raised the possibility that the role of this specific lipid may be conserved across species.
Weaknesses:
This study is not, however, without caveats and several specific conclusions are not fully convincing.
First, the conclusion that GlcT is specifically required in Intestinal Stem Cells (ISCs) is not fully convincing for technical reasons: NRE-Gal4 may be less active in GlcT mutant cells, and the knock-down of GlcT using Dl-Gal4ts may not be restricted to ISCs given the perdurance of Gal4 and of its downstream RNAi.
As previously mentioned, we acknowledge that a role for GlcT in early EBs or EEPs cannot be completely ruled out. We have revised our manuscript to present a more cautious conclusion and explicitly described this possibility in the updated version.
Second, the results from the antibody uptake assays are not clear.: i) the levels of internalized Delta were not quantified in these experiments; ii) additionally, live guts were incubated with anti-Delta for 3hr. This long period of incubation indicated that the observed results may not necessarily reflect the dynamics of endocytosis of antibody-bound Delta, but might also inform about the distribution of intracellular Delta following the internalization of unbound anti-Delta. It would thus be interesting to examine the level of internalized Delta in experiments with shorter incubation time.
We thank the reviewer for these excellent questions. In our antibody uptake experiments, we noted that Dl reached its peak accumulation after a 3-hour incubation period. We recognize that quantifying internalized Dl would enhance our analysis, and we will include the corresponding statistical graphs in the revised version of the manuscript. In addition, we agree that during the 3-hour incubation, the potential internalization of unbound anti-Dl cannot be ruled out, as it may influence the observed distribution of intracellular Dl. We therefore attempted to supplement our findings with live imaging experiments to investigate the dynamics of Dl/Notch endocytosis in both normal and GlcT mutant ISCs. However, we found that the GFP expression level of Dl-GFP (either in the knock-in or transgenic line) was too low to be reliably tracked. During the three-hour observation period, the weak GFP signal remained largely unchanged regardless of the GlcT mutation status, and the signal resolution under the microscope was insufficient to clearly distinguish membrane-associated from intracellular Dl. Therefore, we were unable to obtain a dynamic view of Dl trafficking through live imaging. Nevertheless, our Dl antibody uptake and endosomal retention analyses collectively support the notion that MacCer influences Notch signaling by regulating Dl endocytosis.
Overall, the proposed working model needs to be solidified as important questions remain open, including: is the endo-lysosomal system, i.e. steady-state distribution of endo-lysosomal markers, affected by the Mac-Cer deficiency? Is the trafficking of Notch also affected by the Mac-Cer deficiency? is the rate of Delta endocytosis also affected by the Mac-Cer deficiency? are the levels of cell-surface Delta reduced upon the loss of Mac-Cer?
Regarding the impact on the endo-lysosomal system, this is indeed an important aspect to explore. While we did not conduct experiments specifically designed to evaluate the steady-state distribution of endo-lysosomal markers, our analyses utilizing Rab5-GFP overexpression and Rab7 staining did not indicate any significant differences in endosome distribution in MacCer deficient conditions. Moreover, we still observed high expression of the NRE-LacZ reporter specifically at the boundaries of clones in GlcT mutant cells (Fig. 4A), indicating that GlcT mutant EBs remain responsive to Dl produced by normal ISCs located right at the clone boundary. Therefore, we propose that MacCer deficiency may specifically affect Dl trafficking without impacting Notch trafficking.
In our 3-hour antibody uptake experiments, we observed a notable decrease in cell-surface Dl, which was accompanied by an increase in intracellular accumulation. These findings collectively suggest that Dl may be unstable on the cell surface, leading to its accumulation in early endosomes.
Third, while the mouse results are potentially interesting, they seem to be relatively preliminary, and future studies are needed to test whether the level of Notch receptor activation is reduced in this model.
In the mouse small intestine, Olfm4 is a well-established target gene of the Notch signaling pathway, and its staining provides a reliable indication of Notch pathway activation. While we attempted to evaluate Notch activation using additional markers, such as Hes1 and NICD, we encountered difficulties, as the corresponding antibody reagents did not perform well in our hands. Despite these challenges, we believe that our findings with Olfm4 provide an important start point for further investigation in the future.
Reviewer #3 (Public review):
Summary:
In this paper, Tang et al report the discovery of a Glycoslyceramide synthase gene, GlcT, which they found in a genetic screen for mutations that generate tumorous growth of stem cells in the gut of Drosophila. The screen was expertly done using a classic mutagenesis/mosaic method. Their initial characterization of the GlcT alleles, which generate endocrine tumors much like mutations in the Notch signaling pathway, is also very nice. Tang et al checked other enzymes in the glycosylceramide pathway and found that the loss of one gene just downstream of GlcT (Egh) gives similar phenotypes to GlcT, whereas three genes further downstream do not replicate the phenotype. Remarkably, dietary supplementation with a predicted GlcT/Egh product, Lactosyl-ceramide, was able to substantially rescue the GlcT mutant phenotype. Based on the phenotypic similarity of the GlcT and Notch phenotypes, the authors show that activated Notch is epistatic to GlcT mutations, suppressing the endocrine tumor phenotype and that GlcT mutant clones have reduced Notch signaling activity. Up to this point, the results are all clear, interesting, and significant. Tang et al then go on to investigate how GlcT mutations might affect Notch signaling, and present results suggesting that GlcT mutation might impair the normal endocytic trafficking of Delta, the Notch ligand. These results (Fig X-XX), unfortunately, are less than convincing; either more conclusive data should be brought to support the Delta trafficking model, or the authors should limit their conclusions regarding how GlcT loss impairs Notch signaling. Given the results shown, it's clear that GlcT affects EE cell differentiation, but whether this is via directly altering Dl/N signaling is not so clear, and other mechanisms could be involved. Overall the paper is an interesting, novel study, but it lacks somewhat in providing mechanistic insight. With conscientious revisions, this could be addressed. We list below specific points that Tang et al should consider as they revise their paper.
Strengths:
The genetic screen is excellent.
The basic characterization of GlcT phenotypes is excellent, as is the downstream pathway analysis.
Weaknesses:
(1) Lines 147-149, Figure 2E: here, the study would benefit from quantitations of the effects of loss of brn, B4GalNAcTA, and a4GT1, even though they appear negative.
We have incorporated the quantifications for the effects of the loss of brn, B4GalNAcTA, and a4GT1 in the updated Figure 2.
(2) In Figure 3, it would be useful to quantify the effects of LacCer on proliferation. The suppression result is very nice, but only effects on Pros+ cell numbers are shown.
We have now added quantifications of the number of EEs per clone to the updated Figure 3.
(3) In Figure 4A/B we see less NRE-LacZ in GlcT mutant clones. Are the data points in Figure 4B per cell or per clone? Please note. Also, there are clearly a few NRE-LacZ+ cells in the mutant clone. How does this happen if GlcT is required for Dl/N signaling?
In Figure 4B, the data points represent the fluorescence intensity per single cell within each clone. It is true that a few NRE-LacZ+ cells can still be observed within the mutant clone; however, this does not contradict our conclusion. As noted, high expression of the NRE-LacZ reporter was specifically observed around the clone boundaries in MacCer deficient cells (Fig. 4A), indicating that the mutant EBs can normally receive Dl signal from the normal ISCs located at the clone boundary and activate the Notch signaling pathway. Therefore, we believe that, although affecting Dl trafficking, MacCer deficiency does not significantly affect Notch trafficking.
(4) Lines 222-225, Figure 5AB: The authors use the NRE-Gal4ts driver to show that GlcT depletion in EBs has no effect. However, this driver is not activated until well into the process of EB commitment, and RNAi's take several days to work, and so the author's conclusion is "specifically required in ISCs" and not at all in EBs may be erroneous.
As previously mentioned, we acknowledge that a role for GlcT in early EBs or EEPs cannot be completely ruled out. We have revised our manuscript to present a more cautious conclusion and described this possibility in the updated version.
(5) Figure 5C-F: These results relating to Delta endocytosis are not convincing. The data in Fig 5C are not clear and not quantitated, and the data in Figure 5F are so widely scattered that it seems these co-localizations are difficult to measure. The authors should either remove these data, improve them, or soften the conclusions taken from them. Moreover, it is unclear how the experiments tracing Delta internalization (Fig 5C) could actually work. This is because for this method to work, the anti-Dl antibody would have to pass through the visceral muscle before binding Dl on the ISC cell surface. To my knowledge, antibody transcytosis is not a common phenomenon.
We thank the reviewer for these insightful comments and suggestions. In our in vivo experiments, we observed increased co-localization of Rab5 and Dl in GlcT mutant ISCs, indicating that Dl trafficking is delayed at the transition to Rab7⁺ late endosomes, a finding that is further supported by our antibody uptake experiments. We acknowledge that the data presented in Fig. 5C are not fully quantified and that the co-localization data in Fig. 5F may appear somewhat scattered; therefore, we have included additional quantification and enhanced the data presentation in the revised manuscript.
Regarding the concern about antibody internalization, we appreciate this point. We currently do not know if the antibody reaches the cell surface of ISCs by passing through the visceral muscle or via other routes. Given that the experiment was conducted with fragmented gut, it is possible that the antibody may penetrate into the tissue through mechanisms independent of transcytosis.
As mentioned earlier, we attempted to supplement our findings with live imaging experiments to investigate the dynamics of Dl/Notch endocytosis in both normal and GlcT mutant ISCs. However, we found that the GFP expression level of Dl-GFP (either in the knock-in or transgenic line) was too low to be reliably tracked. During the three-hour observation period, the weak GFP signal remained largely unchanged regardless of the GlcT mutation status, and the signal resolution under the microscope was insufficient to clearly distinguish membrane-associated from intracellular Dl. Therefore, we were unable to obtain a dynamic view of Dl trafficking through live imaging. Nevertheless, our Dl antibody uptake and endosomal retention analyses collectively support the notion that MacCer influences Notch signaling by regulating Dl endocytosis.
(6) It is unclear whether MacCer regulates Dl-Notch signaling by modifying Dl directly or by influencing the general endocytic recycling pathway. The authors say they observe increased Dl accumulation in Rab5+ early endosomes but not in Rab7+ late endosomes upon GlcT depletion, suggesting that the recycling endosome pathway, which retrieves Dl back to the cell surface, may be impaired by GlcT loss. To test this, the authors could examine whether recycling endosomes (marked by Rab4 and Rab11) are disrupted in GlcT mutants. Rab11 has been shown to be essential for recycling endosome function in fly ISCs.
We agree that assessing the state of recycling endosomes, especially by using markers such as Rab11, would be valuable in determining whether MacCer regulates Dl-Notch signaling by directly modifying Dl or by influencing the broader endocytic recycling pathway. In the newly added experiments, we found that in GlcT-IR flies, Dl still exhibits partial colocalization with Rab11, and the overall expression pattern of Rab11 is not affected by GlcT knockdown (Fig. 5E-F). These observations suggest that MacCer specifically regulates Dl trafficking rather than broadly affecting the recycling pathway.
(7) It remains unclear whether Dl undergoes post-translational modification by MacCer in the fly gut. At a minimum, the authors should provide biochemical evidence (e.g., Western blot) to determine whether GlcT depletion alters the protein size of Dl.
While we propose that MacCer may function as a component of lipid rafts, facilitating Dl membrane anchorage and endocytosis, we also acknowledge the possibility that MacCer could serve as a substrate for protein modifications of Dl necessary for its proper function. Conducting biochemical analyses to investigate potential post-translational modifications of Dl by MacCer would indeed provide valuable insights. We have performed Western blot analysis to test whether GlcT depletion affects the protein size of Dl. As shown below, we did not detect any apparent changes in the molecular weight of the Dl protein. Therefore, it is unlikely that MacCer regulates post-translational modifications of Dl.
Author response image 1.
To investigate whether MacCer modifies Dl by Western blot,(A) Four lanes were loaded: the first two contained 20 μL of membrane extract (lane 1: GlcT-IR, lane 2: control), while the last two contained 10 μL of membrane extract (B) Full blot images are shown under both long and shortexposure conditions.
(8) It is unfortunate that GlcT doesn't affect Notch signaling in other organs on the fly. This brings into question the Delta trafficking model and the authors should note this. Also, the clonal marker in Figure 6C is not clear.
In the revised working model, we have explicitly described that the events occur in intestinal stem cells. Regarding Figure 6C, we have delineated the clone with a white dashed line to enhance its clarity and visual comprehension.
(9) The authors state that loss of UGCG in the mouse small intestine results in a reduced ISC count. However, in Supplementary Figure C3, Ki67, a marker of ISC proliferation, is significantly increased in UGCG-CKO mice. This contradiction should be clarified. The authors might repeat this experiment using an alternative ISC marker, such as Lgr5.
Previous studies have indicated that dysregulation of the Notch signaling pathway can result in a reduction in the number of ISCs. While we did not perform a direct quantification of ISC numbers in our experiments, our Olfm4 staining—which serves as a reliable marker for ISCs—demonstrates a clear reduction in the number of positive cells in UGCG-CKO mice.
The increased Ki67 signal we observed reflects enhanced proliferation in the transit-amplifying region, and it does not directly indicate an increase in ISC number. Therefore, in UGCG-CKO mice, we observe a decrease in the number of ISCs, while there is an increase in transit-amplifying (TA) cells (progenitor cells). This increase in TA cells is probably a secondary consequence of the loss of barrier function associated with the UGCG knockout.
-
-
www.wikiwand.com www.wikiwand.com
-
Das gerichtliche Aktenzeichen dient der Kennzeichnung eines Dokuments und geht auf die Aktenordnung (AktO) vom 28. November 1934 und ihre Vorgänger zurück.[4]
The court file number is used to identify a document and goes back to the file regulations (AktO) of November 28, 1934 and its predecessors.
The German "file number" (aktenzeichen) is a unique identification of a file, commonly used in their court system and predecessors as well as file numbers in public administration since at least 1934.
Niklas Luhmann studied law at the University of Freiburg from 1946 to 1949, when he obtained a law degree, before beginning a career in Lüneburg's public administration where he stayed in civil service until 1962. Given this fact, it's very likely that Luhmann had in-depth experience with these sorts of file numbers as location identifiers for files and documents.
We know these numbering methods in public administration date back to as early as Vienna, Austria in the 1770s.
The missing piece now is who/where did Luhmann learn his note taking and excerpting practice from? Alberto Cevolini argues that Niklas Luhmann was unaware of the prior tradition of excerpting, though note taking on index cards or slips had been commonplace in academic circles for quite some time and would have been reasonably commonplace during his student years.
Are there handbooks, guides, or manuals in the early 1900's that detail these sorts of note taking practices?
Perhaps something along the lines of Antonin Sertillanges’ book The Intellectual Life (1921) or Paul Chavigny's Organisation du travail intellectuel: recettes pratiques à l’usage des étudiants de toutes les facultés et de tous les travailleurs (in French) (Delagrave, 1918)?
Further recall that Bruno Winck has linked some of the note taking using index cards to legal studies to Roland Claude's 1961 text:
I checked Chavigny’s book on the BNF site. He insists on the use of index cards (‘fiches’), how to index them, one idea per card but not how to connect between the cards and allow navigation between them.
Mind that it’s written in 1919, in Strasbourg (my hometown) just one year after it returned to France. So between students who used this book and Luhmann in Freiburg it’s not far away. My mother taught me how to use cards for my studies back in 1977, I still have the book where she learn the method, as Law student in Strasbourg “Comment se documenter”, by Roland Claude, 1961. Page 25 describes a way to build secondary index to receive all cards relatives to a topic by their number. Still Luhmann system seems easier to maintain but very near.
<small><cite class='h-cite via'>ᔥ <span class='p-author h-card'> Scott P. Scheper </span> in Scott P. Scheper on Twitter: "The origins of the Zettelkasten's numeric-alpha card addresses seem to derive from Niklas Luhmann's early work as a legal clerk. The filing scheme used is called "Aktenzeichen" - See https://t.co/4mQklgSG5u. cc @ChrisAldrich" / Twitter (<time class='dt-published'>06/28/2022 11:29:18</time>)</cite></small>
Link to: - https://hypothes.is/a/Jlnn3IfSEey_-3uboxHsOA - https://hypothes.is/a/4jtT0FqsEeyXFzP-AuDIAA
-
-
www.youtube.com www.youtube.com
-
AWS is 10x slower than a dedicated server for the same price
- Video Title: AWS is 10x slower than a dedicated server for the same price
- Core Argument: Cloud providers, particularly AWS, charge significantly more for base-level compute instances than traditional Virtual Private Server (VPS) providers while delivering substantially less performance. The video argues that horizontal scaling is often unnecessary for 95% of businesses.
- Comparison Setup: The video compared an entry-level AWS instance (EC2 and ECS Fargate) with a similarly specced VPS (1 vCPU, 2 GB RAM) from a popular German provider (Hetzner, referred to as HTNA in the video) using the Sysbench tool.
- AWS EC2 Results: The base EC2 instance cost almost 3 times more than the VPS but delivered poor performance:
- CPU: Approximately 20% of the VPS performance.
- Memory: Only 7.74% of the VPS performance.
- AWS ECS Fargate Results: Using the "serverless" Fargate option, setup was complex and involved many AWS services (ECS, ECR, IAM).
- Cost: The instance was 6 times more expensive than the VPS.
- Performance: Performance improved over EC2 but was still slower and less consistent: 23% (CPU), 80% (Memory), and 84% (File I/O) of the VPS's performance, with fluctuations up to 18%.
- Cost Efficiency: A dedicated VPS server with 4vCPU and 16 GB of RAM was found to be cheaper than the 1 vCPU ECS Fargate task used in the test.
- Conclusion: For a similar price point, a dedicated server is about 10 times faster than an equivalent AWS cloud instance. The video concludes that AWS's dominance is due to its large marketing spend, not superior technical or cost efficiency. A real-world example cited is Lichess, which supports 5.2 million chess games per day on a single dedicated server [00:12:06].
Hacker News Discussion
The discussion was split between criticizing the video's methodology and debating the fundamental value proposition of hyperscale cloud providers versus traditional hosting.
- Criticism of Methodology: Several top comments argued the video was a "low effort 'ha ha AWS sucks' video" with an "AWFUL analysis." Critics suggested the author did not properly configure or understand ECS/Fargate and that comparing the lowest-end shared instances isn't a "proper comparison," which should involve mid-range hardware and careful configuration.
- The Value of AWS Services: Many users defended AWS by stating that customers rarely choose it just for the base EC2 instance price. The true value lies in the managed ecosystem of services like RDS, S3, EKS, ELB, and Cognito, which abstract away operational complexity and allow large customers to negotiate off-list pricing.
- Complexity and Cost Rebuttals: Counter-arguments highlighted that managing AWS complexity often requires hiring expensive "cloud wizards" (Solutions Architects or specialized DevOps staff), shifting the high cost of a SysAdmin team to high cloud management costs. Anecdotes about sudden huge AWS bills and complex debugging were common.
- The "Nobody Gets Fired" Factor: The most common justification for choosing AWS, even at a higher cost, is risk aversion and the avoidance of personal liability. If a core AWS region (like US-East-1) goes down, it's a shared industry failure, but if a self-hosted server fails, the admin is solely responsible for fixing it at 3 a.m.
- Alternative Recommendations: The discussion frequently validated the use of non-hyperscale providers like Hetzner and OVH for significant cost savings and comparable reliability for many non-"cloud native" workloads.
-
-
social-media-ethics-automation.github.io social-media-ethics-automation.github.io
-
19.3. Responses to Meta’s Business Strategies# Let’s look at some responses to Meta’s business plan. 19.3.1. Competition# When Facebook started, there were already other social media platforms in use that Facebook had to compete against, but Facebook became dominant. Since then other companies have tried to compete with Facebook, with different levels of success. Google+ tried to mimic much of what Facebook did, but it got little use and never took off (not enough people to benefit from the network effect). Other social media sites have used more unique features to distinguish themselves from Facebook and get a foothold, such as Twitter with its character limit (forcing short messages, so you can see lots of posts in quick succession), Vine and then TikTok based on short videos, etc. Mastodon [s48] (Fediverse [s49] set of connected social media platforms that it is part of) has a different way of distinguishing itself as a social media network, in that it is an open-source, community-funded social media network (no ads), and hopes people will join to get away from corporate control. Other social media networks have focused on parts of the world where Facebook was less dominant, and so they got a foothold there first, and then spread, like the social media platforms in China (e.g., Sina Weibo, QQ, and TikTok). 19.3.2. Privacy Concerns# Another source of responses to Meta (and similar social media sites), is concern around privacy (especially in relation to surveillance capitalism). The European Union passed the General Data Protection Regulation (GDPR) [s50] law, which forces companies to protect user information in certain ways and give users a “right to be forgotten” [s51] online. Apple also is concerned about privacy, so it introduced app tracking transparency in 2021 [s52]. In response, Facebook says Apple iOS privacy change will result in $10 billion revenue hit this year [s53]. Note that Apple can afford to be concerned with privacy like this because it does not make much money off of behavioral data. Instead, Apple’s profits [s54] are mostly from hardware (e.g., iPhone) and services (e.g., iCloud, Apple Music, Apple TV+).
I find it interesting how Meta’s biggest threat isn’t just other apps, but privacy changes. Platforms like Mastodon show that some users really care about moving away from corporate control, while Apple’s tracking restrictions show that big tech companies can also limit each other. To me, this shows that Meta’s power isn’t absolute — it’s shaped by competition, laws, and other companies, not just by what it wants.
-
-
www.youtube.com www.youtube.com
-
Jakie suplementy diety warto brać jesienią i zimą? Dr Tadeusz Oleszczuk [Sekrety Długowieczności]
- Vitamin D3 (Witamina D3):
- Crucial Supplement: Highly recommended for the autumn/winter season (September to April in Poland) because skin synthesis of D3 is inactive and most people have low levels (safe level is 50-80 ng/mL) [00:00:12], [00:00:33], [00:01:32].
- Benefits: Supports immunity, reduces infection risk, and is vital for hormone production [00:01:17], [00:01:39].
- Actionable Advice: Always check your current level before supplementation, and retest after 3-6 months to ensure the optimal level (50-60 ng/mL) is reached [00:01:24], [00:01:59].
- Omega-3 Fatty Acids (Kwasy omega-3):
- Component: Provides EPA and DHA, which are essential for brain structure (60% fat), nervous system function, and myelin sheaths [00:03:30], [00:03:38].
- Functions: Exhibits anti-inflammatory effects and supports the heart, brain, and overall immunity [00:03:38].
- Storage Tip: Liquid form should be consumed within one month of opening and kept in the refrigerator to prevent oxidation; capsules are more stable [00:03:48], [00:04:01].
- Magnesium (Magnez):
- Role: Helps manage stress, improves memory, and supports muscle function [00:07:05].
- Essential Cofactor: Magnesium is required as a "motor" for the majority of enzymes in the body; deficiency impairs the function of the entire organism [00:07:35], [00:07:42].
- Consumption: Choose easily absorbable and safe forms like magnesium glycinate [00:07:23]. Be mindful that diuretics like coffee and tea can deplete magnesium levels [00:07:46].
- Other Key Supplements:
- Vitamin C and Zinc: Support the immune system and shorten the duration of infections [00:05:03]. It's important to test your zinc level first to avoid harmful excess [00:04:18], [00:04:21].
- Probiotics and Prebiotics: A healthy gut microbiota is the foundation of immunity [00:06:14]. Probiotics need prebiotics (e.g., resistant starch like cold potatoes) to thrive and create beneficial conditions [00:06:24], [00:05:39].
- B Vitamins: A B-complex should be considered if the diet is poor, especially since B12 deficiency can be linked to nervous system issues and stomach problems [00:08:14], [00:08:29].
- General Supplementation Rules:
- Supplements should be individually chosen based on a person's lifestyle and real, confirmed deficiencies [00:09:16], [00:09:21].
- When buying, always check the dosage on the label to ensure the amounts are effective and not just minimal [00:08:44], [00:08:56].
- The foundation of strong immunity remains sleep, diet, and exercise [00:09:26].
- Vitamin D3 (Witamina D3):
-
-
www.youtube.com www.youtube.comYouTube1
-
Yann Braga | Storybook Vitest | ViteConf 2025
What stood out to me is how Storybook keeps everything in one place such as interactions, accessibility checks, and visual regression tests. The speaker mentions that fixing a component instantly updates all the related tests, which makes debugging feel way less chaotic. It’s like the tool encourages good testing habits just by being convenient. It also makes testing feel less like a separate task and more like a natural part of the development workflow. Seeing issues update in real time helps you understand the impact of your changes much faster, which is very good.
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the original reviews
Reviewer #1 (Public review):
Weakness:
I wonder how task difficulty and linguistic labels interact with the current findings. Based on the behavioral data, shapes with more geometric regularities are easier to detect when surrounded by other shapes. Do shape labels that are readily available (e.g., "square") help in making accurate and speedy decisions? Can the sensitivity to geometric regularity in intraparietal and inferior temporal regions be attributed to differences in task difficulty? Similarly, are the MEG oddball detection effects that are modulated by geometric regularity also affected by task difficulty?
We see two aspects to the reviewer’s remarks.
(1) Names for shapes.
On the one hand, is the question of the impact of whether certain shapes have names and others do not in our task. The work presented here is not designed to specifically test the effect of formal western education; however, in previous work (Sablé-Meyer et al., 2021), we noted that the geometric regularity effect remains present even for shapes that do not have specific names, and even in participants who do not have names for them. Thus, we replicated our main effects with both preschoolers and adults that did not attend formal western education and found that our geometric feature model remained predictive of their behavior; we refer the reader to this previous paper for an extensive discussion of the possible role of linguistic labels, and the impact of the statistics of the environment on task performance.
What is more, in our behavior experiments we can discard data from any shape that is has a name in English and run our model comparison again. Doing so diminished the effect size of the geometric feature model, but it remained predictive of human behavior: indeed, if we removed all shapes but kite, rightKite, rustedHinge, hinge and random (i.e., more than half of our data, and shapes for which we came up with names but there are no established names), we nevertheless find that both models significantly correlate with human behavior—see plot in Author response image 1, equivalent of our Fig. 1E with the remaining shapes.
Author response image 1.
An identical analysis on the MEG leads to two noisy but significant clusters (CNN: 64.0ms to 172.0ms; then 192.0ms to 296.0ms; both p<.001: Geometric Features: 312.0ms to 364.0ms with p=.008). We have improved our manuscript thanks to the reviewer’s observation by adding a figure with the new behavior analysis to the supplementary figures and in the result section of the behavior task. We now refer to these analysis where appropriate:
(intro) “The effect appeared as a human universal, present in preschoolers, first-graders, and adults without access to formal western math education (the Himba from Namibia), and thus seemingly independent of education and of the existence of linguistic labels for regular shapes.”
(behavior results) “Finally, to separate the effect of name availability and geometric features on behavior, we replicated our analysis after removing the square, rectangle, trapezoids, rhombus and parallelogram from our data (Fig. S5D). This left us with five shapes, and an RDM with 10 entries, When regressing it in a GLM with our two models, we find that both models are still significant predictors (p<.001). The effect size of the geometric feature model is greatly reduced, yet remained significantly higher than that of the neural network model (p<.001).”
(meg results) “This analysis yielded similar clusters when performed on a subset of shapes that do not have an obvious name in English, as was the case for the behavior analysis (CNN Encoding: 64.0ms to 172.0ms; then 192.0ms to 296.0ms; both p<.001: Geometric Features: 312.0ms to 364.0ms with p=.008).”
(discussion, end of behavior section) “Previously, we only found such a significant mixture of predictors in uneducated humans (whether French preschoolers or adults from the Himba community, mitigating the possible impact of explicit western education, linguistic labels, and statistics of the environment on geometric shape representation) (Sablé-Meyer et al., 2021).”
Perhaps the referee’s point can also be reversed: we provide a normative theory of geometric shape complexity which has the potential to explain why certain shapes have names: instead of seeing shape names as the cause of their simpler mental representation, we suggest that the converse could occur, i.e. the simpler shapes are the ones that are given names.
(2) Task difficulty
On the other hand is the question of whether our effect is driven by task difficulty. First, we would like to point out that this point could apply to the fMRI task, which asks for an explicit detection of deviants, but does not apply to the MEG experiment. In MEG, participants passively looked at sequences of shapes which, for a given block, comprising many instances of a fixed standard shape and rare deviants–even if they notice deviants, they have no task related to them. Yet two independent findings validated the geometric features model: there was a large effect of geometric regularity on the MEG response to deviants, and the MEG dissimilarity matrix between standard shapes correlated with a model based on geometric features, better than with a model based on CNNs. While the response to rare deviants might perhaps be attributed to “difficulty” (assuming that, in spite of the absence of an explicit task, participants try to spot the deviants and find this self-imposed task more difficult in runs with less regular shapes), it seems very hard to explain the representational similarity analysis (RSA) findings based on difficulty. Indeed, what motivated us to use RSA analysis in both fMRI and MEG was to stop relying on the response to deviants, and use solely the data from standard or “reference” shapes, and model their neural response with theory-derived regressors.
We have updated the manuscript in several places to make our view on these points clearer:
(experiment 4) “This design allowed us to study the neural mechanisms of the geometric regularity effect without confounding effects of task, task difficulty, or eye movements.”
(figure 4, legend) “(A) Task structure: participants passively watch a constant stream of geometric shapes, one per second (presentation time 800ms). The stimuli are presented in blocks of 30 identical shapes up to scaling and rotation, with 4 occasional deviant shape. Participants do not have a task to perform beside fixating.”
Reviewer #2 (Public review):
Weakness:
Given that the primary take away from this study is that geometric shape information is found in the dorsal stream, rather than the ventral stream there is very little there is very little discussion of prior work in this area (for reviews, see Freud et al., 2016; Orban, 2011; Xu, 2018). Indeed, there is extensive evidence of shape processing in the dorsal pathway in human adults (Freud, Culham, et al., 2017; Konen & Kastner, 2008; Romei et al., 2011), children (Freud et al., 2019), patients (Freud, Ganel, et al., 2017), and monkeys (Janssen et al., 2008; Sereno & Maunsell, 1998; Van Dromme et al., 2016), as well as the similarity between models and dorsal shape representations (Ayzenberg & Behrmann, 2022; Han & Sereno, 2022).
We thank the reviewer for this opportunity to clarify our writing. We want to use this opportunity to highlight that our primary finding is not about whether the shapes of objects or animals (in general) are processed in the ventral versus or the dorsal pathway, but rather about the much more restricted domain of geometric shapes such as squares and triangles. We propose that simple geometric shapes afford additional levels of mental representation that rely on their geometric features – on top of the typical visual processing. To the best of our knowledge, this point has not been made in the above papers.
Still, we agree that it is useful to better link our proposal to previous ones. We have updated the discussion section titled “Two Visual Pathways” to include more specific references to the literature that have reported visual object representations in the dorsal pathway. Following another reviewer’s observation, we have also updated our analysis to better demonstrate the overlap in activation evoked by math and by geometry in the IPS, as well as include a novel comparison with independently published results.
Overall, to address this point, we (i) show the overlap between our “geometry” contrast (shape > word+tools+houses) and our “math” contrast (number > words); (ii) we display these ROIs side by side with ROIs found in previous work (Amalric and Dehaene, 2016), and (iii) in each math-related ROIs reported in that article, we test our “geometry” (shape > word+tools+houses) contrast and find almost all of them to be significant in both population; see Fig. S5.
Finally, within the ROIs identified with our geometry localizer, we also performed similarity analyses: for each region we extracted the betas of every voxel for every visual category, and estimated the distance (cross-validated mahalanobis) between different visual categories. In both ventral ROIs, in both populations, numbers were closer to shapes than to the other visual categories including text and Chinese characters (all p<.001). In adults, this result also holds for the right ITG (p=.021) and the left IPS (p=.014) but not the right IPS (p=.17). In children, this result did not hold in the areas.
Naturally, overlap in brain activation does not suffice to conclude that the same computational processes are involved. We have added an explicit caveat about this point. Indeed, throughout the article, we have been careful to frame our results in a way that is appropriate given our evidence, e.g. saying “Those areas are similar to those active during number perception, arithmetic, geometric sequences, and the processing of high-level math concepts” and “The IPS areas activated by geometric shapes overlap with those active during the comprehension of elementary as well as advanced mathematical concepts”. We have rephrased the possibly ambiguous “geometric shapes activated math- and number-related areas, particular the right aIPS.” into “geometric shapes activated areas independently found to be activated by math- and number-related tasks, in particular the right aIPS”.
Reviewer #3 (Public review):
Weakness:
Perhaps the manuscript could emphasize that the areas recruited by geometric figures but not objects are spatial, with reduced processing in visual areas. It also seems important to say that the images of real objects are interpreted as representations of 3D objects, as they activate the same visual areas as real objects. By contrast, the images of geometric forms are not interpreted as representations of real objects but rather perhaps as 2D abstractions.
This is an interesting possibility. Geometric shapes are likely to draw attention to spatial dimensions (e.g. length) and to do so in a 2D spatial frame of reference rather than the 3D representations evoked by most other objects or images. However, this possibility would require further work to be thoroughly evaluated, for instance by comparing usual 3D objects with rare instances of 2D ones (e.g. a sheet of paper, a sticker etc). In the absence of such a test, we refrained from further speculation on this point.
The authors use the term "symbolic." That use of that term could usefully be expanded here.
The reviewer is right in pointing out that “symbolic” should have been more clearly defined. We now added in the introduction:
(introduction) “[…] we sometimes refer to this model as “symbolic” because it relies on discrete, exact, rule-based features rather than continuous representations (Sablé-Meyer et al., 2022). In this representational format, geometric shapes are postulated to be represented by symbolic expressions in a “language-of-thought”, e.g. “a square is a four-sided figure with four equal sides and four right angles” or equivalently by a computer-like program from drawing them in a Logo-like language (Sablé-Meyer et al., 2022).”
Here, however, the present experiments do not directly probe this format of a representation. We have therefore simplified our wording and removed many of our use of the word “symbolic” in favor of the more specific “geometric features”.
Pigeons have remarkable visual systems. According to my fallible memory, Herrnstein investigated visual categories in pigeons. They can recognize individual people from fragments of photos, among other feats. I believe pigeons failed at geometric figures and also at cartoon drawings of things they could recognize in photos. This suggests they did not interpret line drawings of objects as representations of objects.
The comparison of geometric abilities across species is an interesting line of research. In the discussion, we briefly mention several lines of research that indicate that non-human primates do not perceive geometric shapes in the same way as we do – but for space reasons, we are reluctant to expand this section to a broader review of other more distant species. The referee is right that there is evidence of pigeons being able to perceive an invariant abstract 3D geometric shape in spite of much variation in viewpoint (Peissig et al., 2019) – but there does not seem to be evidence that they attend to geometric regularities specifically (e.g. squares versus non-squares). Also, the referee’s point bears on the somewhat different issue of whether humans and other animals may recognize the object depicted by a symbolic drawing (e.g. a sketch of a tree). Again, humans seem to be vastly superior in this domain, and research on this topic is currently ongoing in the lab. However, the point that we are making in the present work is specifically about the neural correlates of the representation of simple geometric shapes which by design were not intended to be interpretable as representations of objects.
Categories are established in part by contrast categories; are quadrilaterals, triangles, and circles different categories?
We are not sure how to interpret the referee’s question, since it bears on the definition of “category” (Spontaneous? After training? With what criterion?). While we are not aware of data that can unambiguously answer the reviewer’s question, categorical perception in geometric shapes can be inferred from early work investigating pop-out effects in visual search, e.g. (Treisman and Gormican, 1988): curvature appears to generate strong pop-out effects, and therefore we would expect e.g. circles to indeed be a different category than, say, triangles. Similarly, right angles, as well as parallel lines, have been found to be perceived categorically (Dillon et al., 2019).
This suggests that indeed squares would be perceived as categorically different from triangles and circles. On the other hand, in our own previous work (Sablé-Meyer et al., 2021) we have found that the deviants that we generated from our quadrilaterals did not pop out from displays of reference quadrilaterals. Pop-out is probably not the proper criterion for defining what a “category” is, but this is the extent to which we can provide an answer to the reviewer’s question.
It would be instructive to investigate stimuli that are on a continuum from representational to geometric, e.g., table tops or cartons under various projections, or balls or buildings that are rectangular or triangular. Building parts, inside and out. like corners. Objects differ from geometric forms in many ways: 3D rather than 2D, more complicated shapes, and internal texture. The geometric figures used are flat, 2-D, but much geometry is 3-D (e. g. cubes) with similar abstract features.
We agree that there is a whole line of potential research here. We decided to start by focusing on the simplest set of geometric shapes that would give us enough variation in geometric regularity while being easy to match on other visual features. We agree with the reviewer that our results should hold both for more complex 2-D shapes, but also for 3-D shapes. Indeed, generative theories of shapes in higher dimensions following similar principles as ours have been devised (I. Biederman, 1987; Leyton, 2003). We now mention this in the discussion:
“Finally, this research should ultimately be extended to the representation of 3-dimensional geometric shapes, for which similar symbolic generative models have indeed been proposed (Irving Biederman, 1987; Leyton, 2003).”
The feature space of geometry is more than parallelism and symmetry; angles are important, for example. Listing and testing features would be fascinating. Similarly, looking at younger or preferably non-Western children, as Western children are exposed to shapes in play at early ages.
We agree with the reviewer on all point. While we do not list and test the different properties separately in this work, we would like to highlight that angles are part of our geometric feature model, which includes features of “right-angle” and “equal-angles” as suggested by the reviewer.
We also agree about the importance of testing populations with limited exposure to formal training with geometric shapes. This was in fact a core aspect of a previous article of ours which tests both preschoolers, and adults with no access to formal western education – though no non-Western children (Sablé-Meyer et al., 2021). It remains a challenge to perform brain-imaging studies in non-Western populations (although see Dehaene et al., 2010; Pegado et al., 2014).
What in human experience but not the experience of close primates would drive the abstraction of these geometric properties? It's easy to make a case for elaborate brain processes for recognizing and distinguishing things in the world, shared by many species, but the case for brain areas sensitive to processing geometric figures is harder. The fact that these areas are active in blind mathematicians and that they are parietal areas suggests that what is important is spatial far more than visual. Could these geometric figures and their abstract properties be connected in some way to behavior, perhaps with fabrication and construction as well as use? Or with other interactions with complex objects and environments where symmetry and parallelism (and angles and curvature--and weight and size) would be important? Manual dexterity and fabrication also distinguish humans from great apes (quantitatively, not qualitatively), and action drives both visual and spatial representations of objects and spaces in the brain. I certainly wouldn't expect the authors to add research to this already packed paper, but raising some of the conceptual issues would contribute to the significance of the paper.
We refrained from speculating about this point in the previous version of the article, but share some of the reviewers’ intuitions about the underlying drive for geometric abstraction. As described in (Dehaene, 2026; Sablé-Meyer et al., 2022), our hypothesis, which isn’t tested in the present article, is that the emergence of a pervasive ability to represent aspects of the world as compact expressions in a mental “language-of-thought” is what underlies many domains of specific human competence, including some listed by the reviewer (tool construction, scene understanding) and our domain of study here, geometric shapes.
Recommendations for the Authors:
Reviewer #1 (Recommendations for the authors):
Overall, I enjoyed reading this paper. It is clearly written and nicely showcases the amount of work that has gone into conducting all these experiments and analyzing the data in sophisticated ways. I also thought the figures were great, and I liked the level of organization in the GitHub repository and am looking forward to seeing the shared data on OpenNeuro. I have some specific questions I hope the authors can address.
(1) Behavior
- Looking at Figure 1, it seemed like most shapes are clustering together, whereas square, rectangle, and maybe rhombus and parallelogram are slightly more unique. I was wondering whether the authors could comment on the potential influence of linguistic labels. Is it possible that it is easier to discard the intruder when the shapes are readily nameable versus not?
This is an interesting observation, but the existence of names for shapes does not suffice to explain all of our findings ; see our reply to the public comment.
(2) fMRI
- As mentioned in the public review, I was surprised that the authors went with an intruder task because I would imagine that performance depends on the specific combination of geometric shapes used within a trial. I assume it is much harder to find, for example, a "Right Hinge" embedded within "Hinge" stimuli than a "Right Hinge" amongst "Squares". In addition, the rotation and scaling of each individual item should affect regular shapes less than irregular shapes, creating visual dissimilarities that would presumably make the task harder. Can the authors comment on how we can be sure that the differences we pick up in the parietal areas are not related to task difficulty but are truly related to geometric shape regularities?
Again, please see our public review response for a larger discussion of the impact of task difficulty. There are two aspects to answering this question.
First, the task is not as the reviewer describes: the intruder task is to find a deviant shape within several slightly rotated and scaled versions of the regular shape it came from. During brain imaging, we did not ask participants to find an exemplar of one of our reference shape amidst copies of another, but rather a deviant version of one shape against copies of its reference version. We only used this intruder task with all pairs of shapes to generate the behavioral RSA matrix.
Second, we agree that some of the fMRI effect may stem from task difficulty, and this motivated our use of RSA analysis in fMRI, and a passive MEG task. RSA results cannot be explained by task difficulty.
Overall, we have tried to make the limitations of the fMRI design, and the motivation for turning to passive presentation in MEG, clearer by stating the issues more clearly when we introduce experiment 4:
“The temporal resolution of fMRI does not allow to track the dynamic of mental representations over time. Furthermore, the previous fMRI experiment suffered from several limitations. First, we studied six quadrilaterals only, compared to 11 in our previous behavioral work. Second, we used an explicit intruder detection, which implies that the geometric regularity effect was correlated with task difficulty, and we cannot exclude that this factor alone explains some of the activations in figure 3C (although it is much less clear how task difficulty alone would explain the RSA results in figure 3D). Third, the long display duration, which was necessary for good task performance especially in children, afforded the possibility of eye movements, which were not monitored inside the 3T scanner and again could have affected the activations in figure 3C.”
- How far in the periphery were the stimuli presented? Was eye-tracking data collected for the intruder task? Similar to the point above, I would imagine that a harder trial would result in more eye movements to find the intruder, which could drive some of the differences observed here.
A 1-degree bar was added to Figure 3A, which faithfully illustrates how the stimuli were presented in fMRI. Eye-tracking data was not collected during fMRI. Although the participants were explicitly instructed to fixate at the center of the screen and avoid eye movements, we fully agree with the referee that we cannot exclude that eye movements were present, perhaps more so for more difficult displays, and would therefore have contributed to the observed fMRI activations in experiment 3 (figure 3C). We now mention this limitation explicity at the end of experiment 3. However, crucially, this potential problem cannot apply to the MEG data. During the MEG task, the stimuli were presented one by one at the center of screen, without any explicit task, thus avoiding issues of eye movements. We therefore consider the MEG geometrical regularity effect, which comes at a relatively early latency (starting at ~160 ms) and even in a passive task, to provide the strongest evidence of geometric coding, unaffected by potential eye movement artefacts.
- I was wondering whether the authors would consider showing some un-thresholded maps just to see how widespread the activation of the geometric shapes is across all of the cortex.
We share the uncorrected threshold maps in Fig. S3. for both adults and children in the category localizer, copied here as well. For the geometry task, most of the clusters identified are fairly big and survive cluster-corrected permutations; the uncorrected statistical maps look almost fully identical to the one presented in Fig. 3 (p<.001 map).
- I'm missing some discussion on the role of early visual areas that goes beyond the RSA-CNN comparison. I would imagine that early visual areas are not only engaged due to top-down feedback (line 258) but may actually also encode some of the geometric features, such as parallel lines and symmetry. Is it feasible to look at early visual areas and examine what the similarity structure between different shapes looks like?
If early visual areas encoded the geometric features that we propose, then even early sensor-level RSA matrices should show a strong impact of geometric features similarity, which is not what we find (figure 4D). We do, however, appreciate the referee’s request to examine more closely how this similarity structure looks like. We now provide a movie showing the significant correlation between neural activity and our two models (uncorrected participants); indeed, while the early occipital activity (around 110ms) is dominated by a significant correlation with the CNN model, there are also scattered significant sources associated to the symbolic model around these timepoints already.
To test this further, we used beamformers to reconstruct the source-localized activity in calcarine cortex and performed an RSA analysis across that ROI. We find that indeed the CNN model is strongly significant at t=110ms (t=3.43, df=18, p=.003) while the geometric feature model is not (t=1.04, df=18, p=.31), and the CNN is significantly above the geometric feature model (t=4.25, df=18, p<.001). However, this result is not very stable across time, and there are significant temporal clusters around these timepoints associated to each model, with no significant cluster associated to a CNN > geometric (CNN: significant cluster from 88ms to 140ms, p<.001 in permutation based with 10000 permutations; geometric features has a significant cluster from 80ms to 104ms, p=.0475; no significant cluster on the difference between the two).
(3) MEG
- Similar to the fMRI set, I am a little worried that task difficulty has an effect on the decoding results, as the oddball should pop out more in more geometric shapes, making it easier to detect and easier to decode. Can the authors comment on whether it would matter for the conclusions whether they are decoding varying task difficulty or differences in geometric regularity, or whether they think this can be considered similarly?
See above for an extensive discussion of the task difficulty effect. We point out that there is no task in the MEG data collection part. We have clarified the task design by updating our Fig. 4. Additionally, the fact that oddballs are more perceived more or less easily as a function of their geometric regularity is, in part, exactly the point that we are making – but, in MEG, even in the absence of a task of looking for them.
- The authors discuss that the inflated baseline/onset decoding/regression estimates may occur because the shapes are being repeated within a mini-block, which I think is unlikely given the long ISIs and the fact that the geometric features model is not >0 at onset. I think their second possible explanation, that this may have to do with smoothing, is very possible. In the text, it said that for the non-smoothed result, the CNN encoding correlates with the data from 60ms, which makes a lot more sense. I would like to encourage the authors to provide readers with the unsmoothed beta values instead of the 100-ms smoothed version in the main plot to preserve the reason they chose to use MEG - for high temporal resolution!
We fully agree with the reviewer and have accordingly updated the figures to show the unsmoothed data (see below). Indeed, there is now no significant CNN effect before ~60 ms (up to the accuracy of identifying onsets with our method).
- In Figure 4C, I think it would be useful to either provide error bars or show variability across participants by plotting each participant's beta values. I think it would also be nice to plot the dissimilarity matrices based on the MEG data at select timepoints, just to see what the similarity structure is like.
Following the reviewer’s recommendation, we plot the timeseries with SEM as shaded area, and thicker lines for statistically significant clusters, and we provide the unsmoothed version in figure Fig. 4. As for the dissimilarity matrices at select timepoints, this has now been added to figure Fig. 4.
- To evaluate the source model reconstruction, I think the reader would need a little more detail on how it was done in the main text. How were the lead fields calculated? Which data was used to estimate the sources? How are the models correlated with the source data?
We have imported some of the details in the main text as follows (as well as expanding the methods section a little):
“To understand which brain areas generated these distinct patterns of activations, and probe whether they fit with our previous fMRI results, we performed a source reconstruction of our data. We projected the sensor activity onto each participant's cortical surfaces estimated from T1-images. The projection was performed using eLORETA and emptyroom recordings acquired on the same day to estimate noise covariance, with the default parameters of mne-bids-pipeline. Sources were spaced using a recursively subdivided octahedron (oct5). Group statistics were performed after alignement to fsaverage. We then replicated the RSA analysis […]”
- In addition to fitting the CNN, which is used here to model differences in early visual cortex, have the authors considered looking at their fMRI results and localizing early visual regions, extracting a similarity matrix, and correlating that with the MEG and/or comparing it with the CNN model?
We had ultimately decided against comparing the empirical similarity matrices from the MEG and fMRI experiments, first because the stimuli and tasks are different, and second because this would not be directly relevant to our goal, which is to evaluate whether a geometric-feature model accounts for the data. Thus, we systematically model empirical similarity matrices from fMRI and from MEG with our two models derived from different theories of shape perception in order to test predictions about their spatial and temporal dynamic. As for comparing the similarity matrix from early visual regions in fMRI with that predicted by the CNN model, this is effectively visible from our Fig. 3D where we perform searchlight RSA analysis and modeling with both the CNN and the geometric feature model; bilaterally, we find a correlation with the CNN model, although it sometimes overlap with predictions from the geometric feature model as well. We now include a section explaining this reasoning in appendix:
“Representational similarity analysis also offers a way to directly compared similarity matrices measured in MEG and fMRI, thus allowing for fusion of those two modalities and tentatively assigning a “time stamp” to distinct MRI clusters. However, we did not attempt such an analysis here for several reasons. First, distinct tasks and block structures were used in MEG and fMRI. Second, a smaller list of shapes was used in fMRI, as imposed by the slower modality of acquisition. Third, our study was designed as an attempt to sort out between two models of geometric shape recognition. We therefore focused all analyses on this goal, which could not have been achieved by direct MEG-fMRI fusion, but required correlation with independently obtained model predictions.”
Minor comments
- It's a little unclear from the abstract that there is children's data for fMRI only.
We have reworded the abstract to make this unambiguous
- Figures 4a & b are missing y-labels.
We can see how our labels could be confused with (sub-)plot titles and have moved them to make the interpretation clearer.
- MEG: are the stimuli always shown in the same orientation and size?
They are not, each shape has a random orientation and scaling. On top of a task example at the top of Fig. 4, we have now included a clearer mention of this in the main text when we introduce the task:
“shapes were presented serially, one at a time, with small random changes in rotation and scaling parameters, in miniblocks with a fixed quadrilateral shape and with rare intruders with the bottom right corner shifted by a fixed amount (Sablé-Meyer et al., 2021)”
- To me, the discussion section felt a little lengthy, and I wonder whether it would benefit from being a little more streamlined, focused, and targeted. I found that the structure was a little difficult to follow as it went from describing the result by modality (behavior, fMRI, MEG) back to discussing mostly aspects of the fMRI findings.
We have tried to re-organize and streamline the discussion following these comments.
Then, later on, I found that especially the section on "neurophysiological implementation of geometry" went beyond the focus of the data presented in the paper and was comparatively long and speculative.
We have reexamined the discussion, but the citation of papers emphasizing a representation of non-accidental geometric properties in non-human animals was requested by other commentators on our article; and indeed, we think that they are relevant in the context of our prior suggestion that the composition of geometric features might be a uniquely human feature – these papers suggest that individual features may not, and that it is therefore compositionality which might be special to the human brain. We have nevertheless shortened it.
Furthermore, we think that this section is important because symbolic models are often criticized for lack of a plausible neurophysiological implementation. It is therefore important to discuss whether and how the postulated symbolic geometric code could be realized in neural circuits. We have added this justification to the introduction of this section.
Reviewer #2 (Recommendations for the authors):
(1) If the authors want to specifically claim that their findings align with mathematical reasoning, they could at least show the overlap between the activation maps of the current study and those from prior work.
This was added to the fMRI results. See our answers to the public review.
(2) I wonder if the reason the authors only found aIPS in their first analysis (Figure 2) is because they are contrasting geometric shapes with figures that also have geometric properties. In other words, faces, objects, and houses also contain geometric shape information, and so the authors may have essentially contrasted out other areas that are sensitive to these features. One indication that this may be the case is that the geometric regularity effect and searchlight RSA (Figure 3) contains both anterior and posterior IPS regions (but crucially, little ventral activity). It might be interesting to discuss the implications of these differences.
Indeed, we cannot exclude that the few symmetries, perpendicularity and parallelism cues that can be presented in faces, objects or houses were processed as such, perhaps within the ventral pathway, and that these representations would have been subtracted out. We emphasize that our subtraction isolates the geometrical features that are present in simple regular geometric shapes, over and above those that might exist in other categories. We have added this point to the discussion:
“[… ] For instance, faces possess a plane of quasi-symmetry, and so do many other man-made tools and houses. Thus, our subtraction isolated the geometrical features that are present in simple regular geometric shapes (e.g. parallels, right angles, equality of length) over and above those that might already exist, in a less pure form, in other categories.”
(3) I had a few questions regarding the MEG results.
a. I didn't quite understand the task. What is a regular or oddball shape in this context? It's not clear what is being decoded. Perhaps a small example of the MEG task in Figure 4 would help?
We now include an additional sub-figure in Fig. 4 to explain the paradigm. In brief: there is no explicit task, participants are simply asked to fixate. The shapes come in miniblocks of 30 identical reference shapes (up to rotation and scaling), among which some occasional deviant shapes randomly appear (created by moving the corner of the reference shape by some amount).
b. In Figure 4A/B they describe the correlation with a 'symbolic model'. Is this the same as the geometric model in 4C?
It is. We have removed this ambiguity by calling it “geometric model” and setting its color to the one associated to this model thought the article.
c. The author's explanation for why geometric feature coding was slower than CNN encoding doesn't quite make sense to me. As an explanation, they suggest that previous studies computed "elementary features of location or motor affordance", whereas their study work examines "high-level mathematical information of an abstract nature." However, looking at the studies the authors cite in this section, it seems that these studies also examined the time course of shape processing in the dorsal pathway, not "elementary features of location or motor affordance." Second, it's not clear how the geometric feature model reflects high-level mathematical information (see point above about claiming this is related to math).
We thank the referee for pointing out this inappropriate phrase, which we removed. We rephrased the rest of the paragraph to clarify our hypothesis in the following way:
“However, in this work, we specifically probed the processing of geometric shapes that, if our hypothesis is correct, are represented as mental expressions that combine geometrical and arithmetic features of an abstract categorical nature, for instance representing “four equal sides” or “four right angles”. It seems logical that such expressions, combining number, angle and length information, take more time to be computed than the first wave of feedforward processing within the occipito-temporal visual pathway, and therefore only activate thereafter.”
One explanation may be that the authors' geometric shapes require finer-grained discrimination than the object categories used in prior studies. i.e., the odd-ball task may be more of a fine-grained visual discrimination task. Indeed, it may not be a surprise that one can decode the difference between, say, a hammer and a butterfly faster than two kinds of quadrilaterals.
We do not disagree with this intuition, although note that we do not have data on this point (we are reporting and modelling the MEG RSA matrix across geometric shapes only – in this part, no other shapes such as tools or faces are involved). Still, the difference between squares, rectangles, parallelograms and other geometric shapes in our stimuli is not so subtle. Furthermore, CNNs do make very fine grained distinctions, for instance between many different breeds of dogs in the IMAGENET corpus. Still, those sorts of distinctions capture the initial part of the MEG response, while the geometric model is needed only for the later part. Thus, we think that it is a genuine finding that geometric computations associated with the dorsal parietal pathway are slower than the image analysis performed by the ventral occipito-temporal pathway.
d. CNN encoding at time 0 is a little weird, but the author's explanation, that this is explained by the fact that temporal smoothed using a 100 ms window makes sense. However, smoothing by 100 ms is quite a lot, and it doesn't seem accurate to present continuous time course data when the decoding or RSA result at each time point reflects a 100 ms bin. It may be more accurate to simply show unsmoothed data. I'm less convinced by the explanation about shape prediction.
We agree. Following the reviewer’s advice, as well as the recommendation from reviewer 1, we now display unsmoothed plots, and the effects now exhibit a more reasonable timing (Figure 4D), with effects starting around ~60 ms for CNN encoding.
(4) I appreciate the author's use of multiple models and their explanation for why DINOv2 explains more variance than the geometric and CNN models (that it represents both types of features. A variance partitioning analysis may help strengthen this conclusion (Bonner & Epstein, 2018; Lescroart et al., 2015).
However, one difference between DINOv2 and the CNN used here is that it is trained on a dataset of 142 million images vs. the 1.5 million images used in ImageNet. Thus, DINOv2 is more likely to have been exposed to simple geometric shapes during training, whereas standard ImageNet trained models are not. Indeed, prior work has shown that lesioning line drawing-like images from such datasets drastically impairs the performance of large models (Mayilvahanan et al., 2024). Thus, it is unlikely that the use of a transformer architecture explains the performance of DINOv2. The authors could include an ImageNet-trained transformer (e.g., ViT) and a CNN trained on large datasets (e.g., ResNet trained on the Open Clip dataset) to test these possibilities. However, I think it's also sufficient to discuss visual experience as a possible explanation for the CNN and DINOv2 results. Indeed, young children are exposed to geometric shapes, whereas ImageNet-trained CNNs are not.
We agree with the reviewer’s observation. In fact, new and ongoing work from the lab is also exploring this; we have included in supplementary materials exactly what the reviewer is suggesting, namely the time course of the correlation with ViT and with ConvNeXT. In line with the reviewers’ prediction, these networks, trained on much larger dataset and with many more parameters, can also fit the human data as well as DINOv2. We ran additional analysis of the MEG data with ViT and ConvNeXT, which we now report in Fig. S6 as well as in an additional sentence in that section:
“[…] similar results were obtained by performing the same analysis, not only with another vision transformer network, ViT, but crucially using a much larger convolutional neural network, ConvNeXT, which comprises ~800M parameters and has been trained on 2B images, likely including many geometric shapes and human drawings. For the sake of completeness, RSA analysis in sensor space of the MEG data with these two models is provided in Fig. S6.”
We conclude that the size and nature of the training set could be as important as the architecture – but also note that humans do not rely on such a huge training set. We have updated the text, as well as Fig. S6, accordingly by updating the section now entitled “Vision Transformers and Larger Neural Networks”, and the discussion section on theoretical models.
(5) The authors may be interested in a recent paper from Arcaro and colleagues that showed that the parietal cortex is greatly expanded in humans (including infants) compared to non-human primates (Meyer et al., 2025), which may explain the stronger geometric reasoning abilities of humans.
A very interesting article indeed! We have updated our article to incorporate this reference in the discussion, in the section on visual pathways, as follows:
“Finally, recent work shows that within the visual cortex, the strongest relative difference in growth between human and non-human primates is localized in parietal areas (Meyer et al., 2025). If this expansion reflected the acquisition of new processing abilities in these regions, it might explain the observed differences in geometric abilities between human and non-human primates (Sablé-Meyer et al., 2021).”
Also, the authors may want to include this paper, which uses a similar oddity task and compelling shows that crows are sensitive to geometric regularity:
Schmidbauer, P., Hahn, M., & Nieder, A. (2025). Crows recognize geometric regularity. Science Advances, 11(15), eadt3718. https://doi.org/10.1126/sciadv.adt3718
We have ongoing discussions with the authors of this work and are have prepared a response to their findings (Sablé-Meyer and Dehaene, 2025)–ultimately, we think that this discussion, which we agree is important, does not have its place in the present article. They used a reduced version of our design, with amplified differences in the intruders. While they did not test the fit of their model with CNN or geometric feature models, we did and found that a simple CNN suffices to account for crow behavior. Thus, we disagree that their conclusions follow from their results and their conclusions. But the present article does not seem to be the right platform to engage in this discussion.
References
Ayzenberg, V., & Behrmann, M. (2022). The Dorsal Visual Pathway Represents Object-Centered Spatial Relations for Object Recognition. The Journal of Neuroscience, 42(23), 4693-4710. https://doi.org/10.1523/jneurosci.2257-21.2022
Bonner, M. F., & Epstein, R. A. (2018). Computational mechanisms underlying cortical responses to the affordance properties of visual scenes. PLoS Computational Biology, 14(4), e1006111. https://doi.org/10.1371/journal.pcbi.1006111
Bueti, D., & Walsh, V. (2009). The parietal cortex and the representation of time, space, number and other magnitudes. Philosophical Transactions of the Royal Society B: Biological Sciences, 364(1525), 1831-1840.
Dehaene, S., & Brannon, E. (2011). Space, time and number in the brain: Searching for the foundations of mathematical thought. Academic Press.
Freud, E., Culham, J. C., Plaut, D. C., & Bermann, M. (2017). The large-scale organization of shape processing in the ventral and dorsal pathways. eLife, 6, e27576.
Freud, E., Ganel, T., Shelef, I., Hammer, M. D., Avidan, G., & Behrmann, M. (2017). Three-dimensional representations of objects in dorsal cortex are dissociable from those in ventral cortex. Cerebral Cortex, 27(1), 422-434.
Freud, E., Plaut, D. C., & Behrmann, M. (2016). 'What 'is happening in the dorsal visual pathway. Trends in Cognitive Sciences, 20(10), 773-784.
Freud, E., Plaut, D. C., & Behrmann, M. (2019). Protracted developmental trajectory of shape processing along the two visual pathways. Journal of Cognitive Neuroscience, 31(10), 1589-1597.
Han, Z., & Sereno, A. (2022). Modeling the Ventral and Dorsal Cortical Visual Pathways Using Artificial Neural Networks. Neural Computation, 34(1), 138-171. https://doi.org/10.1162/neco_a_01456
Janssen, P., Srivastava, S., Ombelet, S., & Orban, G. A. (2008). Coding of shape and position in macaque lateral intraparietal area. Journal of Neuroscience, 28(26), 6679-6690.
Konen, C. S., & Kastner, S. (2008). Two hierarchically organized neural systems for object information in human visual cortex. Nature Neuroscience, 11(2), 224-231.
Lescroart, M. D., Stansbury, D. E., & Gallant, J. L. (2015). Fourier power, subjective distance, and object categories all provide plausible models of BOLD responses in scene-selective visual areas. Frontiers in Computational Neuroscience, 9(135), 1-20. https://doi.org/10.3389/fncom.2015.00135
Mayilvahanan, P., Zimmermann, R. S., Wiedemer, T., Rusak, E., Juhos, A., Bethge, M., & Brendel, W. (2024). In search of forgotten domain generalization. arXiv Preprint arXiv:2410.08258.
Meyer, E. E., Martynek, M., Kastner, S., Livingstone, M. S., & Arcaro, M. J. (2025). Expansion of a conserved architecture drives the evolution of the primate visual cortex. Proceedings of the National Academy of Sciences, 122(3), e2421585122. https://doi.org/10.1073/pnas.2421585122
Orban, G. A. (2011). The extraction of 3D shape in the visual system of human and nonhuman primates. Annual Review of Neuroscience, 34, 361-388.
Romei, V., Driver, J., Schyns, P. G., & Thut, G. (2011). Rhythmic TMS over Parietal Cortex Links Distinct Brain Frequencies to Global versus Local Visual Processing. Current Biology, 21(4), 334-337. https://doi.org/10.1016/j.cub.2011.01.035
Sereno, A. B., & Maunsell, J. H. R. (1998). Shape selectivity in primate lateral intraparietal cortex. Nature, 395(6701), 500-503. https://doi.org/10.1038/26752
Summerfield, C., Luyckx, F., & Sheahan, H. (2020). Structure learning and the posterior parietal cortex. Progress in Neurobiology, 184, 101717. https://doi.org/10.1016/j.pneurobio.2019.101717
Van Dromme, I. C., Premereur, E., Verhoef, B.-E., Vanduffel, W., & Janssen, P. (2016). Posterior Parietal Cortex Drives Inferotemporal Activations During Three-Dimensional Object Vision. PLoS Biology, 14(4), e1002445. https://doi.org/10.1371/journal.pbio.1002445
Xu, Y. (2018). A tale of two visual systems: Invariant and adaptive visual information representations in the primate brain. Annu. Rev. Vis. Sci, 4, 311-336.
Reviewer #3 (Recommendations for the authors):
Bring into the discussion some of the issues outlined above, especially a) the spatial rather than visual of the geometric figures and b) the non-representational aspects of geometric form aspects.
We thank the reviewer for their recommendations – see our response to the public review for more details.
-
-
osf.io osf.io
-
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public review):
Summary:
This paper presents two experiments, both of which use a target detection paradigm to investigate the speed of statistical learning. The first experiment is a replication of Batterink, 2017, in which participants are presented with streams of uniform-length, trisyllabic nonsense words and asked to detect a target syllable. The results replicate previous findings, showing that learning (in the form of response time facilitation to later-occurring syllables within a nonsense word) occurs after a single exposure to a word. In the second experiment, participants are presented with streams of variable-length nonsense words (two trisyllabic words and two disyllabic words) and perform the same task. A similar facilitation effect was observed as in Experiment 1. The authors interpret these findings as evidence that target detection requires mechanisms different from segmentation. They present results of a computational model to simulate results from the target detection task and find that an "anticipation mechanism" can produce facilitation effects, without performing segmentation. The authors conclude that the mechanisms involved in the target detection task are different from those involved in the word segmentation task.
Strengths:
The paper presents multiple experiments that provide internal replication of a key experimental finding, in which response times are facilitated after a single exposure to an embedded pseudoword. Both experimental data and results from a computational model are presented, providing converging approaches for understanding and interpreting the main results. The data are analyzed very thoroughly using mixed effects models with multiple explanatory factors.
Weaknesses:
In my view, the main weaknesses of this study relate to the theoretical interpretation of the results.
(1) The key conclusion from these findings is that the facilitation effect observed in the target detection paradigm is driven by a different mechanism (or mechanisms) than those involved in word segmentation. The argument here I think is somewhat unclear and weak, for several reasons:
First, there appears to be some blurring in what exactly is meant by the term "segmentation" with some confusion between segmentation as a concept and segmentation as a paradigm.
Conceptually, segmentation refers to the segmenting of continuous speech into words. However, this conceptual understanding of segmentation (as a theoretical mechanism) is not necessarily what is directly measured by "traditional" studies of statistical learning, which typically (at least in adults) involve exposure to a continuous speech stream followed by a forced-choice recognition task of words versus recombined foil items (part-words or nonwords). To take the example provided by the authors, a participant presented with the sequence GHIABCDEFABCGHI may endorse ABC as being more familiar than BCG, because ABC is presented more frequently together and the learned association between A and B is stronger than between C and G. However, endorsement of ABC over BCG does not necessarily mean that the participant has "segmented" ABC from the speech stream, just as faster reaction times in responding to syllable C versus A do not necessarily indicate successful segmentation. As the authors argue on page 7, "an encounter to a sequence in which two elements co-occur (say, AB) would theoretically allow the learner to use the predictive relationship during a subsequent encounter (that A predicts B)." By the same logic, encoding the relationship between A and B could also allow for the above-chance endorsement of items that contain AB over items containing a weaker relationship.
Both recognition performance and facilitation through target detection reflect different outcomes of statistical learning. While they may reflect different aspects of the learning process and/or dissociable forms of memory, they may best be viewed as measures of statistical learning, rather than mechanisms in and of themselves.
Thanks for this nuanced discussion, and this is an important point that R2 also raised. We agree that segmentation can refer to both an experimental paradigm and a mechanism that accounts for learning in the experimental paradigm. In the experimental paradigm, participants are asked to identify which words they believe to be (whole) words from the continuous syllable stream. In the target-detection experimental paradigm, participants are not asked to identify words from continuous streams, and instead, they respond to the occurrences of a certain syllable. It’s possible that learners employ one mechanism in these two tasks, or that they employ separate mechanisms. It’s also the case that, if all we have is positive evidence for both experimental paradigms, i.e., learners can succeed in segmentation tasks as well as in target detection tasks with different types of sequences, we would have no way of talking about different mechanisms, as you correctly suggested that evidence for segmenting AB and processing B faster following A, is not evidence for different mechanisms.
However, that is not the case. When the syllable sequences contain same-length subsequences (i.e., words), learning is indeed successful in both segmentation and target detection tasks. However, in studies such as Hoch et al. (2013), findings suggest that words from mixed-length sequences are harder to segment than words from uniform-length sequences. This finding exists in adult work (e.g., Hoch et al. 2013) as well as infant work (Johnson & Tyler, 2010), and replicated here in the newly included Experiment 3, which stands in contrast to the positive findings of the facilitation effect with mixed-length sequences in the target detection paradigm (one of our main findings in the paper). Thus, it seems to be difficult to explain, if the learning mechanisms were to be the same, why humans can succeed in mixed-length sequences in target detection (as shown in Experiment 2) but fail in uniform-length sequences (as shown in Hoch et al. and Experiment 3).
In our paper, we have clarified these points describe the separate mechanisms in more detail, in both the Introduction and General Discussion sections.
(2) The key manipulation between experiments 1 and 2 is the length of the words in the syllable sequences, with words either constant in length (experiment 1) or mixed in length (experiment 2). The authors show that similar facilitation levels are observed across this manipulation in the current experiments. By contrast, they argue that previous findings have found that performance is impaired for mixed-length conditions compared to fixed-length conditions. Thus, a central aspect of the theoretical interpretation of the results rests on prior evidence suggesting that statistical learning is impaired in mixed-length conditions. However, it is not clear how strong this prior evidence is. There is only one published paper cited by the authors - the paper by Hoch and colleagues - that supports this conclusion in adults (other mentioned studies are all in infants, which use very different measures of learning). Other papers not cited by the authors do suggest that statistical learning can occur to stimuli of mixed lengths (Thiessen et al., 2005, using infant-directed speech; Frank et al., 2010 in adults). I think this theoretical argument would be much stronger if the dissociation between recognition and facilitation through RTs as a function of word length variability was demonstrated within the same experiment and ideally within the same group of participants.
To summarize the evidence of learning uniform-length and mixed-length sequences (which we discussed in the Introduction section), “even though infants and adults alike have shown success segmenting syllable sequences consisting of words that were uniform in length (i.e., all words were either disyllabic; Graf Estes et al., 2007; or trisyllabic, Aslin et al., 1998), both infants and adults have shown difficulty with syllable sequences consisting of words of mixed length (Johnson & Tyler, 2010; Johnson & Jusczyk, 2003a; 2003b; Hoch et al., 2013).” The newly added Experiment 3 also provided evidence for the difference in uniform-length and mixed-length sequences. Notably, we do not agree with the idea that infant work should be disregarded as evidence just because infants were tested with habituation methods; not only were the original findings (Saffran et al. 1996) based on infant work, so were many other studies on statistical learning.
There are other segmentation studies in the literature that have used mixed-length sequences, which are worth discussing. In short, these studies differ from the Saffran et al. (1996) studies in many important ways, and in our view, these differences explain why the learning was successful. Of interest, Thiessen et al. (2005) that you mentioned was based on infant work with infant methods, and demonstrated the very point we argued for: In their study, infants failed to learn when mixed-length sequences were pronounced as adult-directed speech, and succeeded in learning given infant-directed speech, which contained prosodic cues that were much more pronounced. The fact that infants failed to segment mixed-length sequences without certain prosodic cues is consistent with our claim that mixed-length sequences are difficult to segment in a segmentation paradigm. Another such study is Frank et al. (2010), where continuous sequences were presented in “sentences”. Different numbers of words were concatenated into sentences where a 500ms break was present between each sentence in the training sequence. One sentence contained only one word, or two words, and in the longest sentence, there were 24 words. The results showed that participants are sensitive to the effect of sentence boundaries, which coincide with word boundaries. In the extreme, the one-word-per-sentence condition simply presents learners with segmented word forms. In the 24-word-per-sentence condition, there are nevertheless sentence boundaries that are word boundaries, and knowing these word boundaries alone should allow learners to perform above chance in the test phase. Thus, in our view, this demonstrates that learners can use sentence boundaries to infer word boundaries, which is an interesting finding in its own right, but this does not show that a continuous syllable sequence with mixed word lengths is learnable without additional information. In summary, to our knowledge, syllable sequences containing mixed word lengths are better learned when additional cues to word boundaries are present, and there is strong evidence that syllable sequences containing uniform-word lengths are learned better than mixed-length ones.
Frank, M. C., Goldwater, S., Griffiths, T. L., & Tenenbaum, J. B. (2010). Modeling human performance in statistical word segmentation. Cognition, 117(2), 107-125.
To address your proposal of running more experiments to provide stronger evidence for our theory, we were planning to run another study to have the same group of participants do both the segmentation and target detection paradigm as suggested, but we were unable to do so as we encountered difficulties to run English-speaking participants. Instead, we have included an experiment (now Experiment 3), showing the difference between the learning of uniform-length and mixed-length sequences with the segmentation paradigm that we have never published previously. This experiment provides further evidence for adults’ difficulties in segmenting mixed-length sequences.
(3) The authors argue for an "anticipation" mechanism in explaining the facilitation effect observed in the experiments. The term anticipation would generally be understood to imply some kind of active prediction process, related to generating the representation of an upcoming stimulus prior to its occurrence. However, the computational model proposed by the authors (page 24) does not encode anything related to anticipation per se. While it demonstrates facilitation based on prior occurrences of a stimulus, that facilitation does not necessarily depend on active anticipation of the stimulus. It is not clear that it is necessary to invoke the concept of anticipation to explain the results, or indeed that there is any evidence in the current study for anticipation, as opposed to just general facilitation due to associative learning.
Thanks for raising this point. Indeed, the anticipation effect we reported is indistinguishable from the facilitation effect that we reported in the reported experiments. We have dropped this framing.
In addition, related to the model, given that only bigrams are stored in the model, could the authors clarify how the model is able to account for the additional facilitation at the 3rd position of a trigram compared to the 2nd position?
Thanks for the question. We believe it is an empirical question whether there is an additional facilitation at the 3rd position of a trigram compared to the 2nd position. To investigate this issue, we conducted the following analysis with data from Experiment 1. First, we combined the data from two conditions (exact/conceptual) from Experiment 1 so as to have better statistical power. Next, we ran a mixed effect regression with data from syllable positions 2 and 3 only (i.e., data from syllable position 1 were not included). The fixed effect included the two-way interaction between syllable position and presentation, as well as stream position, and the random effect was a by-subject random intercept and stream position as the random slope. This interaction was significant (χ<sup>2</sup>(3) =11.73, p=0.008), suggesting that there is additional facilitation to the 3rd position compared to the 2nd position.
For the model, here is an explanation of why the model assumes an additional facilitation to the 3rd position. In our model, we proposed a simple recursive relation between the RT of a syllable occurring for the nth time and the n+1<sup>th</sup> time, which is:
and
RT(1) = RT0 + stream_pos * stream_inc, where the n in RT(n) represents the RT for the n<sup>th</sup> presentation of the target syllable, stream_pos is the position (3-46) in the stream, and occurrence is the number of occurrences that the syllable has occurred so far in the stream.
What this means is that the model basically provides an RT value for every syllable in the stream. Thus, for a target at syllable position 1, there is a RT value as an unpredictable target, and for targets at syllable position 2, there is a facilitation effect. For targets at syllable position 3, it is facilitated the same amount. As such, there is an additional facilitation effect for syllable position 3 because effects of predication are recursive.
(4) In the discussion of transitional probabilities (page 31), the authors suggest that "a single exposure does provide information about the transitions within the single exposure, and the probability of B given A can indeed be calculated from a single occurrence of AB." Although this may be technically true in that a calculation for a single exposure is possible from this formula, it is not consistent with the conceptual framework for calculating transitional probabilities, as first introduced by Saffran and colleagues. For example, Saffran et al. (1996, Science) describe that "over a corpus of speech there are measurable statistical regularities that distinguish recurring sound sequences that comprise words from the more accidental sound sequences that occur across word boundaries. Within a language, the transitional probability from one sound to the next will generally be highest when the two sounds follow one another within a word, whereas transitional probabilities spanning a word boundary will be relatively low." This makes it clear that the computation of transitional probabilities (i.e., Y | X) is conceptualized to reflect the frequency of XY / frequency of X, over a given language inventory, not just a single pair. Phrased another way, a single exposure to pair AB would not provide a reliable estimate of the raw frequencies with which A and AB occur across a given sample of language.
Thanks for the discussion. We understand your argument, but we respectively disagree that computing transitional probabilities must be conducted under a certain theoretical framework. In our humble opinion, computing transitional probabilities is a mathematical operation, and as such, it is possible to do so with the least amount of data possible that enables the mathematical operation, which concretely is a single exposure during learning. While it is true that a single exposure may not provide a reliable estimate of frequencies or probabilities, it does provide information with which the learner can make decisions.
This is particularly true for topics under discussion regarding the minimal amount of exposure that can enable learning. It is important to distinguish the following two questions: whether learners can learn from a short exposure period (from a single exposure, in fact) and how long of an exposure period does the learner require for it to be considered to produce a reliable estimate of frequencies. Incidentally, given the fact that learners can learn from a single exposure based on Batterink (2017) and the current study, it does not appear that learners require a long exposure period to learn about transitional probabilities.
(5) In experiment 2, the authors argue that there is robust facilitation for trisyllabic and disyllabic words alike. I am not sure about the strength of the evidence for this claim, as it appears that there are some conflicting results relevant to this conclusion. Notably, in the regression model for disyllabic words, the omnibus interaction between word presentation and syllable position did not reach significance (p= 0.089). At face value, this result indicates that there was no significant facilitation for disyllabic words. The additional pairwise comparisons are thus not justified given the lack of omnibus interaction. The finding that there is no significant interaction between word presentation, word position, and word length is taken to support the idea that there is no difference between the two types of words, but could also be due to a lack of power, especially given the p-value (p = 0.010).
Thanks for the comment. Firstly, we believe there is a typo in your comment, where in the last sentence, we believe you were referring to the p-value of 0.103 (source: “The interaction was not significant (χ2(3) = 6.19, p= 0.103”). Yes, a null result with a frequentist approach cannot support a null claim, but Bayesian analyses could potentially provide evidence for the null.
To this end, we conducted a Bayes factor analysis using the approach outlined in Harms and Lakens (2018), which generates a Bayes factor by computing a Bayesian information criterion for a null model and an alternative model. The alternative model contained a three-way interaction of word length, word presentation, and word position, whereas the null model contained a two-way interaction between word presentation and word position as well as a main effect of word length. Thus, the two models only differ in terms of whether there is a three-way interaction. The Bayes factor is then computed as exp[(BICalt − BICnull)/2]. This analysis showed that there is strong evidence for the null, where the Bayes Factor was found to be exp(25.65) which is more than 1011. Thus, there is no power issue here, and there is strong evidence for the null claim that word length did not interact with other factors in Experiment 2.
There is another issue that you mentioned, of whether we should conduct pairwise comparisons if the omnibus interaction did not reach significance. This would be true given the original analysis plan, but we believe that a revised analysis plan makes more sense. In the revised analysis plan for Experiment 2, we start with the three-way interaction (as just described in the last paragraph). The three-way interaction was not significant, and after dropping the third interaction terms, the two-way interaction and the main effect of word length are both significant, and we use this as the overall model. Testing the significance of the omnibus interaction between presentation and syllable position, we found that this was significant (χ<sup>2</sup>(3) =49.77, p<0.001). This represents that, in one model, that the interaction between presentation and syllable position using data from both disyllabic and trisyllabic words. This was in addition to a significant fixed effect of word length (β=0.018, z=6.19, p<0.001). This should motivate the rest of the planned analysis, which regards pairwise comparisons in different word length conditions.
(6) The results plotted in Figure 2 seem to suggest that RTs to the first syllable of a trisyllabic item slow down with additional word presentations, while RTs to the final position speed up. If anything, in this figure, the magnitude of the effect seems to be greater for 1st syllable positions (e.g., the RT difference between presentation 1 and 4 for syllable position 1 seems to be numerically larger than for syllable position 3, Figure 2D). Thus, it was quite surprising to see in the results (p. 16) that RTs for syllable position 1 were not significantly different for presentation 1 vs. the later presentations (but that they were significant for positions 2 and 3 given the same comparison). Is this possibly a power issue? Would there be a significant slowdown to 1st syllables if results from both the exact replication and conceptual replication conditions were combined in the same analysis?
Thanks for the suggestion and your careful visual inspection of the data. After combining the data, the slowdown to 1st syllables is indeed significant. We have reported this in the results of Experiment 1 (with an acknowledgement to this review):
Results showed that later presentations took significantly longer to respond to compared to the first presentation (χ<sup>2</sup>(3) = 10.70, p=0.014), where the effect grew larger with each presentation (second presentation: β=0.011, z=1.82, p=0.069; third presentation: β=0.019, z=2.40, p=0.016; fourth presentation: β=0.034, z=3.23, p=0.001).
(7) It is difficult to evaluate the description of the PARSER simulation on page 36. Perhaps this simulation should be introduced earlier in the methods and results rather than in the discussion only.
Thanks for the suggestions. We have added two separate simulations in the paper, which should describe the PARSER simulations sufficiently, as well as provide further information on the correspondence between the simulations and the experiments. Thanks again for the great review! We believe our paper has improved significantly as a result.
Tags
Annotators
URL
-
-
social-media-ethics-automation.github.io social-media-ethics-automation.github.io
-
Parents post these videos online, where viewers are intended to laugh at the distress, despair, and sense of betrayal the children express.
It seems that only videos or posts with shaming and can make the audience laugh would grab more attention and have more exposure on the internet. I want to say that watching people criticize or shame someone is a natural behavior among humans, while the “Three Character Classic" suggests that people are kind by nature. Therefore, this remains the question that is our kindness been taught or it's just our natural beings
-
-
www.biorxiv.org www.biorxiv.org
-
Note: This response was posted by the corresponding author to Review Commons. The content has not been altered except for formatting.
Learn more at Review Commons
Reply to the reviewers
Revision Plan
Manuscript number: RC-2025-03208
Corresponding author(s): Jared Nordman
[The "revision plan" should delineate the revisions that authors intend to carry out in response to the points raised by the referees. It also provides the authors with the opportunity to explain their view of the paper and of the referee reports.
- *
The document is important for the editors of affiliate journals when they make a first decision on the transferred manuscript. It will also be useful to readers of the reprint and help them to obtain a balanced view of the paper.
- *
If you wish to submit a full revision, please use our "Full Revision" template. It is important to use the appropriate template to clearly inform the editors of your intentions.]
1. General Statements [optional]
All three reviewers of our manuscript were very positive about our work. The reviewers noted that our work represents a necessary advance that is timely, addresses important issues in the chromatin field, and will of broad interest to this community. Given the nature of our work and the positive reviews, we feel that this manuscript would best be suited for the Journal of Cell Biology.
2. Description of the planned revisions
Reviewer #1 (Evidence, reproducibility and clarity (Required)):
Summary:
The authors investigate the function of the H3 chaperone NASP, which is known to bind directly to H3 and prevent degradation of soluble H3. What is unclear is where NASP functions in the cell (nucleus or cytoplasm), how NASP protects H3 from degradation (direct or indirect), and if NASP affects H3 dynamics (nuclear import or export). They use the powerful model system of Drosophila embryos because the soluble H3 pool is high due to maternal deposition and they make use of photoconvertable Dendra-tagged proteins, since these are maternally deposited and can be used to measure nuclear import/export rates.
Using these systems and tools, they conclude that NASP affects nuclear import, but only indirectly, because embryos from NASP mutant mothers start out with 50% of the maternally deposited H3. Because of the depleted H3 and reduced import rates, NASP deficient embryos also have reduced nucleoplasmic and chromatin-associated H3. Using a new Dendra-tagged NASP allele, the authors show that NASP and H3 have different nuclear import rates, indicating that NASP is not a chaperone that shuttles H3 into the nucleus. They test H3 levels in embryos that have no nuclei and conclude that NASP functions in the cytoplasm, and through protein aggregation assays they conclude that NASP prevents H3 aggregation.
Major comments:
The text was easy to read and logical. The data are well presented, methods are complete, and statistics are robust. The conclusions are largely reasonable. However, I am having trouble connecting the conclusions in text to the data presented in Figure 4.
First, I'm confused why the conclusion from Figure 4A is that NASP functions in the cytoplasm of the egg. Couldn't NASP be required in the ovary (in, say, nurse cell nuclei) to stimulate H3 expression and deposition into the egg? The results in 4A would look the same if the mothers deposit 50% of the normal H3 into the egg. Why is NASP functioning specifically in the cytoplasm when it is also so clearly imported into the nucleus? Maybe NASP functions wherever it is, and by preventing nuclear import, you force it to function in the cytoplasm. I do not have additional suggestions for experiments, but I think the authors need to be very clear about the different interpretations of these data and to discuss WHY they believe their conclusion is strongest.
The concern raised by the reviewer regarding NASP function during oogenesis has been addressed in a previous work published from our lab. Unfortunately, we did not do a good job conveying this work in the original version of this manuscript. We demonstrated that total H3 levels are unaffected when comparing WT and NASP mutant stage 14 egg chambers. This means that the amount of H3 deposited into the eggs does not change in the absence of NASP. To address the reviewer's comment, we will change the text to make the link to our previous work clear.
Second, an alternate conclusion from Figure 4D/E is that mothers are depositing less H3 protein into the egg, but the same total amount is being aggregated. This amount of aggregated protein remains constant in activated eggs, but additional H3 translation leads to more total H3? The authors mention that additional translation can compensate for reduced histone pools (line 416).
Similar to our response above, the total amount of H3 in wild type and NASP mutant stage 14 egg chambers is the same. Therefore, mothers are depositing equal amounts of H3 into the egg. We will make the necessary changes in the text to make this point clear.
As the function of NASP in the cytoplasm (when it clearly imports into the nucleus) and role in H3 aggregation are major conclusions of the work, the authors need to present alternative conclusions in the text or complete additional experiments to support the claims. Again, I do not have additional suggestions for experiments, but I think the authors need to be very clear about the different interpretations of these data and to discuss WHY they believe their conclusion is strongest.
A common issue raised by all three reviewers was to more convincingly demonstrate that assay that we have used to isolate protein aggregates does, in fact, isolate protein aggregates. To verify this, we will be performing the aggregate isolation assay using controls that are known to induce more protein aggregation. We will perform the aggregation assay with egg chambers or extracts that are exposed to heat shock or the aggregation-inducing chemicals Canavanine and Azetidine-2-carboxylic acid. The chemical treatment was a welcome suggestion from reviewer #3. These experiments will significantly strengthen any claims based on the outcome of the aggregation assay.
We will also make changes to the text and include other interpretations of our work as the reviewer has suggested.
Data presentation:
Overall, I suggest moving some of the supplemental figures to the main text, adding representative movie stills to show where the quantitative data originated, and moving the H3.3 data to the supplement. Not because it's not interesting, but because H3.3 and H3.2 are behaving the same.
Where possible, we will make changes to the figure display to improve the logic and flow of the manuscript
Fig 1:
It would strengthen the figure to include representative still images that led to the quantitative data, mostly so readers understand how the data were collected.
We will add representative stills to Figure 1 to help readers understand how the data is collected. We will also a representative H3-Dendra movie similar to the NASP supplemental movie.
The inclusion of a "simulated 50% H3" in panel C is confusing. Why?
We used a 50% reduction in H3 levels because that is reduction in H3 we measure in embryos laid by NASP-mutant mothers in our previous work. A reduction in H3 levels alone would be predicted to change the nuclear import rate of H3. Thus, having a quantitative model of H3 import kinetics was key in our understanding of NASP function in vivo. We will revise the text to make this clear.
I would also consider normalizing the data between A and B (and C and D) by dividing NASP/WT. This could be included in the supplement (OPTIONAL)
We can normalize the values and include the data in a supplemental figure.
Fig S1:
The data simulation S1G should be moved to the main text, since it is the primary reason the authors reject the hypothesis that NASP influences H3 import rates.
This is a good point. We will move S1G into the Figure 1.
Fig 2:
Once again, I think it would help to include a few representative images of the photoconverted Dendra2 in the main text.
We will add representative images of the photoconversion in Figure 2.
I struggled with A/B, I think due to not knowing how the data were normalized. When I realized that the WT and NASP data are not normalized to each other, but that the NASP values are likely starting less than the WT values, it made way more sense. I suggest switching the order of data presentation so that C-F are presented first to establish that there is less chromatin-bound H3 in the first place, and then present A/B to show no change in nuclear export of the H3 that is present, allowing the conclusion of both less soluble AND chromatin-bound H3.
The order of the presentation of the data was to test if NASP was acting as a nuclear receptor. Since Figure 1 compares the nuclear import, we wanted to address the nuclear export and provide a comprehensive analysis of the role of NASP in H3 nuclear dynamics before advancing on to other consequences of NASP depletion. We can add the graphs with the un-normalized values in the Supplemental Figure to show the actual difference in total intensity values.
Fig S2:
If M1-M3 indicate males, why are the ovaries also derived from males? I think this is just confusing labeling.
We will change the labelling.
Supplemental Movie S1:
Beautiful. Would help to add a time stamp (OPTIONAL).
Thank you! We will add the time stamp to the movie
Fig 3:
Panel C is the same as Fig S1A (not Fig 1A, as is said in the legend), though I appreciate the authors pointing it out in the legend. Also see line 276.
We appreciate the reviewer for pointing this out. We will make the change in the text to correct this.
Panel D is a little confusing, because presumably the "% decrease in import rate" cannot be positive (Y axis). This could be displayed as a scatter (not bar) as in Panels B/C (right) where the top of the Y axis is set to 0.
We understand the reviewer's concern that the decrease value cannot be positive. We can adjust the y-axis so that it caps off at 0.
Fig S3:
A: What do the different panels represent? I originally thought developmental time, but now I think just different representative images? Are these age-matched from time at egg lay?
The different panels show representative images. We can clarify that in the figure legend.
C: What does "embryos" mean? Same question for Fig 4A.
In this figure, embryos mean the exact number of embryos used to form the lysate for the western blot. We will clarify this in the figure legend.
Fig 4:
A: What does "embryos" mean? Number of embryos? Age in hours?
In this figure, embryos mean the exact number of embryos used to form the lysate for the western blot. We will clarify this in the figure legend.
C: Not sure the workflow figure panel is necessary, as I can't tell what each step does. This is better explained in methods. However I appreciated the short explanation in the text (lines 314-5).
The workflow panel helps to identify the samples labelled as input and aggregate for the western blot analysis. Since our input in the western blots does not refer to the total protein lysate, we feel it is helpful to point out exactly what stage at the protocol we are utilizing the sample for our analysis.
Minor comments:
The authors should describe the nature of the NASP alleles in the main text and present evidence of robust NASP depletion, potentially both in ovaries and in embryos. The antibody works well for westerns (Fig S2B). This is sort of demonstrated later in Figure 4A, but only in NAAP x twine activated eggs.
We appreciate the reviewer's comments about the NASP mutant allele. In our previous publication, we characterized the NASP mutant fly line and its effect on both stage 14 egg chambers and the embryos. We will emphasize the reference to our previous work in the text.
Lines 163, 251, 339: minor typos
Line 184: It would help to clarify- I'm assuming cytoplasmic concentration (or overall) rather than nuclear concentration. If nuclear, I'd expect the opposite relationship. This occurs again when discussing NASP (line 267). I suspect it's also not absolute concentration, but relative concentration difference between cytoplasm and nucleus. It would help clarify if the authors were more precise.
We appreciate the reviewer's point and will add the clarification in the text.
Line 189: Given that the "established integrative model" helps to reject the hypothesis that NASP is involved in H3 import, I think it's important to describe the model a little more, even though it's previously published.
We will add few sentences giving a brief description of the model to the text.
Line 203: "The measured rate of H3.2 export from the nucleus is negligible" clarify this is in WT situations and not a conclusion from this study.
We will add the clarification of this statement in the text.
Line 211: How can the authors be so sure that the decrease in WT is due to "the loss of non-chromatin bound nucleoplasmic H3.2-Dendra2?"
From the live imaging experiments, the H3.2-Dendra2 intensity in the nucleus reduces dramatically upon nuclear envelope breakdown with the only H3.2-Dendra2 intensity remaining being the chromatin bound H3.2. Excess H3.2 is imported into the nucleus and not all of it is incorporated into the chromatin. This is a unique feature of the embryo system that has been observed previously. We mention that the intensity reduction is due to the loss of non-chromatin bound nucleoplasmic H3.2.
Line 217: In the conclusion, the authors indicate that NASP indirectly affects soluble supply of H3 in the nucleoplasm. I do believe they've shown that the import rate effect is indirect, but I don't know why they conclude that the effect of NASP on the soluble nucleoplasmic H3 supply is indirect. Similarly, the conclusion is indirect on line 239. Yet, the authors have not shown it's not direct, just assumed since NASP results in 50% decrease to deposited maternal histones.
We appreciate the feedback on the conclusions of Figure 2 from the reviewer. Our conclusions are primarily based on the effect of H3 levels in the absence of NASP in the early embryos. To establish direct causal effects, it would be important to recover the phenotypes by complementation experiments and providing molecular interactions to cause the effects. In this study we have not established those specific details to make conclusions of direct effects. We will change the text to make this more clear.
Line 292: What is the nature of the NASP "mutant?" Is it a null? Similarly, what kind of "mutant" is the twine allele? Line 295.
We will include descriptions of the NASP and twine mutants in the text.
Line 316: Why did the authors use stage 14 egg chambers here when they previously used embryos? This becomes more clear later shortly, when the authors examine activated eggs, but it's confusing in text.
The reason to use stage 14 egg chambers was to establish NASP function during oogenesis. We will modify the text to emphasize the reason behind using stage 14 egg chambers.
Lines 343-348: It's unclear if the authors are drawing extended conclusions here or if they are drawing from prior literature (if so, citations would be required). For example, why during oogenesis/embryogenesis are aggregation and degradation developmentally separated?
This conclusion is based primarily based on the findings from this study (Figure 4) and out previous published work. We will modify the text for more clarity.
Lines 386-7: I do not understand why the authors conclude that H3 aggregation and degradation are "developmentally uncoupled" and why, in the absence of NASP, "H3 aggregation precedes degradation."
This is based data in Figure 4 combined with our previous working showing that the total level of H3 in not changed in NASP-mutant stage 14 egg chambers. Aggregates seem to be more persistent in the stage 14 egg chambers (oogenesis) and they get cleared out upon egg activation (entry into embryogenesis). This provides evidence for aggregation occurring prior to degradation and these two events occurring in different developmental stages. We will change the text to make this more clear.
Line 395: Why suddenly propose that NASP also functions in the nucleus to prevent aggregation, when earlier the authors suggest it functions only in the cytoplasm?
We will make the necessary edits to ensure that the results don't suggest a role of NASP exclusive to the cytoplasm. Our findings highlight a cytoplasmic function of NASP, however, we do not want to rule out that this same function couldn't occur in the nucleus.
Lines 409-413: The authors claim that histone deficiency likely does not cause the embryonic arrest seen in embryos from NASP mutant mothers. This is because H3 is reduced by 50% yet some embryos arrest long before they've depleted this supply. However, the authors also showed that H3 import rates are affected in these embryos due to lower H3 concentration. Since the early embryo cycles are so rapid, reduced H3 import rates could lead to early arrest, even though available H3 remains in the cytoplasm.
We thank the reviewer for their suggestion. This conclusion is based on the findings from the previous study from our lab which showed that the majority of the embryos laid by NASP mutant females get arrested in the very early nuclear cycles (Reviewer #1 (Significance (Required)):
The significance of the work is conceptual, as NASP is known to function in H3 availability but the precise mechanism is elusive. This work represents a necessary advance, especially to show that NASP does not affect H3 import rates, nor does it chaperone H3 into the nucleus. However, the authors acknowledge that many questions remain. Foremost, why is NASP imported into the nucleus and what is its role there?
I believe this work will be of interest to those who focus on early animal development, but NASP may also represent a tool, as the authors conclude in their discussion, to reduce histone levels during development and examine nucleosome positioning. This may be of interest to those who work on chromatin accessibility and zygotic genome activation.
I am a genetics expert who works in Drosophila embryogenesis. I do not have the expertise to evaluate the aggregate methods presented in Figure 4.
Reviewer #2 (Evidence, reproducibility and clarity (Required)):
Summary:
This manuscript focuses on the role of the histone chaperone NASP in Drosophila. NASP is a chaperone specific to histone H3 that is conserved in mammals. Many aspects of the molecular mechanisms by which NASP selectively binds histone H3 have been revealed through biochemical studies. However, key aspects of NASP's in vivo roles remain unclear, including where in the cell NASP functions, and how it prevents H3 degradation. Through live imaging in the early Drosophila embryo, which possesses large amounts of soluble H3 protein, Das et al determine that NASP does not control nuclear import or export of H3.2 or H3.3. Instead, they find through differential centrifugation analysis that NASP functions in the cytoplasm to prevent H3 aggregation and hence its subsequent degradation.
Major Comments:
The protein aggregation assays raise several questions. From a technical standpoint, it would be helpful to have a positive control to demonstrate that the assay is effective at detecting protein aggregates. Ie. a genotype that exhibits increased protein aggregation; this could be for a protein besides H3. A common issue raised by all three reviewers was to more convincingly demonstrate that assay that we have used to isolate protein aggregates does, in fact, isolate protein aggregates. To verify this, we will be performing the aggregate isolation assay using controls that are known to induce more protein aggregation. We will perform the aggregation assay with egg chambers or extracts that are exposed to heat shock or the aggregation-inducing chemicals Canavanine and Azetidine-2-carboxylic acid. The chemical treatment was a welcome suggestion from reviewer #3. These experiments will significantly strengthen any claims based on the outcome of the aggregation assay.
If NASP is not required to prevent H3 degradation in egg chambers, then why are H3 levels much lower in NASP input lanes relative to wild-type egg chambers in Fig 4D? We appreciate the reviewer's inputs regarding the reduced H3 levels in the NASP mutant egg chambers. We observe this reduction in H3 levels in the input because of the altered solubility of H3 which leads to the loss of H3 protein at different steps of the aggregate isolation assay. We will add a supplement figure showing H3 levels at different steps of the aggregate isolation assay. We do want to stress, however, that the total levels of H3 in stage 14 egg chambers does not change between WT and the NASP mutant.
A corollary to this is that the increased fraction of H3 in aggregates in NASP mutants seems to be entirely due to the reduction in total H3 levels rather than an increase in aggregated H3. If NASP's role is to prevent aggregation in the cytoplasm, and degradation has not yet begun in egg chambers, then why are aggregated H3 levels not increased in NASP mutants relative to wild-type egg chambers? If the same number of egg chambers were used, shouldn't the total amount of histone be the same in the absence of degradation?
In previously published work, we demonstrated that total H3 levels are unaffected when comparing WT and NASPmutant stage 14 egg chambers. This means that the amount of H3 deposited into the eggs does not change in the absence of NASP. To address the reviewer's comment, we will change the text to make the link to our previous work clear. As stated above, we will add a supplement figure showing H3 levels at different steps of the aggregate isolation assay.
The live imaging studies are well designed, executed, and quantified. They use an established genotype (H3.2-Dendra2) in wild-type and NASP maternal mutants to demonstrate that NASP is not directly involved in nuclear import of H3.2. Decreased import is likely due to reduced H3.2 levels in NASP mutants rather than reduced import rates per se. The same methodology was used to determine that loss of NASP did not affect H3.2 nuclear export. These findings eliminate H3.2 nuclear import/export regulation as possible roles for NASP, which had been previously proposed.
Thank you.
Live imaging also conclusively demonstrates that the levels of H3.2 in the nucleoplasm and in mitotic chromatin are significantly lower in NASP mutants than wild-type nuclei. Despite these lower histone levels, the nuclear cycle duration is only modestly lengthened. The live imagining of NASP-Dendra2 nuclear import conclusively demonstrate that NASP and H3.2 are unlikely to be imported into the nucleus as one complex.
Thank you.
Minor Comments:
Additional details on how the NASP-Dendra2 CRISPR allele was generated should be provided. In addition, additional details on how it was determined that this allele is functional should be provided (e.g. quantitative assays for fertility/embryo viability of NASP-Dendra2 females) We will make these additions to the text.
If statistical tests are used to determine significance, the type of test used should be reported in the figure legends throughout.
We will make the addition of the statistical tests to the figure legends.
The western blot shown in Figure 4A looks more like a 4-fold reduction in H3 levels in NASP mutants relative to wild-type embryos, rather than the quantified 2-fold reduction. Perhaps a more representative blot can be shown.
We have additional blots in the supplemental figure S3C. The quantification was performed after normalization to the total protein levels and we can highlight that in the figure legend.
Reviewer #2 (Significance (Required)):
As a fly chromatin biologist with colleagues that utilize mammalian experimental systems, I feel this manuscript will be of broad interest to the chromatin research community. Packaging of the genome into chromatin affects nearly every DNA-templated process, making the mechanisms by which histone proteins are expressed, chaperoned, and deposited into chromatin of high importance to the field. The study has multiple strengths, including high-quality quantitative imaging, use of a terrific experimental system (storage and deposition of soluble histones in early fly embryos). The study also answers outstanding questions in the field, specifically that NASP does not control nuclear import/export of histone H3. Instead, the authors propose that NASP functions to prevent protein aggregation. If this could be conclusively demonstrated, it would be valuable to the field. However, the protein aggregation studies need improvement. Technical demonstration that their differential centrifugation assay accurately detects aggregated proteins is needed. Further, NASP mutants do not exhibit increased H3 protein aggregation in the data presented. Instead, the increased fraction of aggregated H3 in NASP mutants seems to be due to a reduction in the overall levels of H3 protein, which is contrary to the model presented in this paper.
Reviewer #3 (Evidence, reproducibility and clarity (Required)):
This manuscript by Das et al. entitled "NASP functions in the cytoplasm to prevent histone H3 aggregation during early embryogenesis", explores the role of the histone chaperone NASP in regulating histone H3 dynamics during early Drosophila embryogenesis. Using primarily live imaging approaches, the authors found that NASP is not directly involved in the import or export of H3. Moreover, the authors claimed that NASP prevents H3 aggregation rather than protects against degradation.
Major Comments:
Figure 1A-B: The plotted data appear to have substantial dispersion. Could the authors include individual data points or provide representative images to help the reader assess variability?
We chose to show unnormalized data in Figure 1 so readers could better compare the actual import values of H3 in the presence and absence of NASP. We felt it was a better representation of the true biological difference although raw data is more dispersive. We did also include normalized data in the supplement. Regardless, we will add representative stills to Figure 1 and include a H3-Dendra2 movie in the supplement to show the representative data.
Given that the authors conclude that the reduced nuclear import is due to lowered H3 levels in NASP-deficient embryos, would overexpression of H3 rescue this phenotype? This would directly test whether H3 levels, rather than import machinery per se, drive the effect.
We thank the reviewer for their valuable suggestion. We and others have tried to overexpress histones in the Drosophila early embryo without success. There must be an undefined feedback mechanism preventing histone overexpression in the germline. In fact, a recent paper has been deposited on bioRxiv (https://doi.org/10.1101/2024.12.23.630206) that suggest H4 protein could provide a feedback mechanism to prevent histone overexpression. While we would love to do this experiment, it is not technically feasible at this time.
Figure 2A-B: The authors present the Relative Intensity of H3-Dendra2, but this metric obscures absolute differences between Control and NASP knockout embryos. Please include Total Intensity plots to show the actual reduction in H3 levels.
We will add the total H3-Dendra2 intensity plots to the supplemental figure for the export curves.
Additionally, Western blot analysis of nucleoplasmic H3 from wild-type vs. NASP-deficient embryos would provide essential biochemical confirmation of H3 level reductions.
We will measure nuclear H3 levels by western from 0-2 hr embryos laid by WT and NASP mutant flies.
Figure 4: To support the conclusion that NASP prevents H3 aggregation, I recommend performing aggregation assays by adding compounds that induce unfolding (amino acid analogues that induce unfolding, like canavanine or Azetidine-2-carboxylic acid) or using aggregation-prone H3 mutants.
This is a very helpful suggestion! It is difficult to get chemicals into Drosophila eggs, but we will treat extracts directly with these chemicals. Additionally, we will use heat shocked eggs and extracts as an additional control.
Inclusion of CMA and proteasome inhibition experiments could also clarify whether degradation pathways are secondarily involved or compensatory in the absence of NASP.
The degradation pathway for H3 in the absence of NASP is unknown and a major focus of our future work is to define this pathway. Drosophila does not have a CMA pathway and therefore, we don't know how H3 aggregates are being sensed.
Minor Comments:
(1) The Introduction would benefit from mentioning the two NASP isoforms that exist in mammals (sNASP and tNASP), as this evolutionary context may inform interpretation of the Drosophila results.
We will make the edits in the text to include that Drosophila NASP is the sole homolog of sNASP and that tNASP ortholog is not found in Drosophila.
(2) Could the authors comment on the status of histone H4 in their experimental system? Given the observed cytoplasmic pool of H3, is it likely to exist as a monomer? If this H3 pool is monomeric, does that suggest an early failure in H3-H4 dimerization, and could this contribute to its aggregation propensity?
In our previous work we noted that NASP binds more preferentially to H3 and the levels of H3 we much more reduced upon NASP depletion than H4. We pointed out in this publication that our data was consistent with H3 stores being monomeric in the Drosophila embryo. We don't' have a H4-Dendra2 line to test. In the future, however, this is something we are very keen to look at.
Reviewer #3 (Significance (Required)):
This work addresses a timely and important question in the field of chromatin biology and developmental epigenetics. The focus on histone homeostasis during embryogenesis and the cytoplasmic role of NASP adds a novel perspective. The live imaging experiments are a clear strength, providing valuable spatiotemporal insights. However, I believe that the manuscript would benefit significantly from additional biochemical validation to support and clarify some of the mechanistic claims.
3. Description of the revisions that have already been incorporated in the transferred manuscript
- *
4. Description of analyses that authors prefer not to carry out
Please include a point-by-point response explaining why some of the requested data or additional analyses might not be necessary or cannot be provided within the scope of a revision. This can be due to time or resource limitations or in case of disagreement about the necessity of such additional data given the scope of the study. Please leave empty if not applicable.
-
Note: This preprint has been reviewed by subject experts for Review Commons. Content has not been altered except for formatting.
Learn more at Review Commons
Referee #1
Evidence, reproducibility and clarity
Summary:
The authors investigate the function of the H3 chaperone NASP, which is known to bind directly to H3 and prevent degradation of soluble H3. What is unclear is where NASP functions in the cell (nucleus or cytoplasm), how NASP protects H3 from degradation (direct or indirect), and if NASP affects H3 dynamics (nuclear import or export). They use the powerful model system of Drosophila embryos because the soluble H3 pool is high due to maternal deposition and they make use of photoconvertable Dendra-tagged proteins, since these are maternally deposited and can be used to measure nuclear import/export rates.
Using these systems and tools, they conclude that NASP affects nuclear import, but only indirectly, because embryos from NASP mutant mothers start out with 50% of the maternally deposited H3. Because of the depleted H3 and reduced import rates, NASP deficient embryos also have reduced nucleoplasmic and chromatin-associated H3. Using a new Dendra-tagged NASP allele, the authors show that NASP and H3 have different nuclear import rates, indicating that NASP is not a chaperone that shuttles H3 into the nucleus. They test H3 levels in embryos that have no nuclei and conclude that NASP functions in the cytoplasm, and through protein aggregation assays they conclude that NASP prevents H3 aggregation.
Major comments:
The text was easy to read and logical. The data are well presented, methods are complete, and statistics are robust. The conclusions are largely reasonable. However, I am having trouble connecting the conclusions in text to the data presented in Figure 4.
First, I'm confused why the conclusion from Figure 4A is that NASP functions in the cytoplasm of the egg. Couldn't NASP be required in the ovary (in, say, nurse cell nuclei) to stimulate H3 expression and deposition into the egg? The results in 4A would look the same if the mothers deposit 50% of the normal H3 into the egg. Why is NASP functioning specifically in the cytoplasm when it is also so clearly imported into the nucleus? Maybe NASP functions wherever it is, and by preventing nuclear import, you force it to function in the cytoplasm. I do not have additional suggestions for experiments, but I think the authors need to be very clear about the different interpretations of these data and to discuss WHY they believe their conclusion is strongest.
Second, an alternate conclusion from Figure 4D/E is that mothers are depositing less H3 protein into the egg, but the same total amount is being aggregated. This amount of aggregated protein remains constant in activated eggs, but additional H3 translation leads to more total H3? The authors mention that additional translation can compensate for reduced histone pools (line 416).
As the function of NASP in the cytoplasm (when it clearly imports into the nucleus) and role in H3 aggregation are major conclusions of the work, the authors need to present alternative conclusions in the text or complete additional experiments to support the claims. Again, I do not have additional suggestions for experiments, but I think the authors need to be very clear about the different interpretations of these data and to discuss WHY they believe their conclusion is strongest.
Data presentation:
Overall, I suggest moving some of the supplemental figures to the main text, adding representative movie stills to show where the quantitative data originated, and moving the H3.3 data to the supplement. Not because it's not interesting, but because H3.3 and H3.2 are behaving the same.
Fig 1:
It would strengthen the figure to include representative still images that led to the quantitative data, mostly so readers understand how the data were collected. The inclusion of a "simulated 50% H3" in panel C is confusing. Why? I would also consider normalizing the data between A and B (and C and D) by dividing NASP/WT. This could be included in the supplement (OPTIONAL)
Fig S1:
The data simulation S1G should be moved to the main text, since it is the primary reason the authors reject the hypothesis that NASP influences H3 import rates.
Fig 2:
Once again, I think it would help to include a few representative images of the photoconverted Dendra2 in the main text. I struggled with A/B, I think due to not knowing how the data were normalized. When I realized that the WT and NASP data are not normalized to each other, but that the NASP values are likely starting less than the WT values, it made way more sense. I suggest switching the order of data presentation so that C-F are presented first to establish that there is less chromatin-bound H3 in the first place, and then present A/B to show no change in nuclear export of the H3 that is present, allowing the conclusion of both less soluble AND chromatin-bound H3.
Fig S2:
If M1-M3 indicate males, why are the ovaries also derived from males? I think this is just confusing labeling. Supplemental Movie S1: Beautiful. Would help to add a time stamp (OPTIONAL).
Fig 3:
Panel C is the same as Fig S1A (not Fig 1A, as is said in the legend), though I appreciate the authors pointing it out in the legend. Also see line 276. Panel D is a little confusing, because presumably the "% decrease in import rate" cannot be positive (Y axis). This could be displayed as a scatter (not bar) as in Panels B/C (right) where the top of the Y axis is set to 0.
Fig S3:
A: What do the different panels represent? I originally thought developmental time, but now I think just different representative images? Are these age-matched from time at egg lay? C: What does "embryos" mean? Same question for Fig 4A. Fig 4: A: What does "embryos" mean? Number of embryos? Age in hours? C: Not sure the workflow figure panel is necessary, as I can't tell what each step does. This is better explained in methods. However I appreciated the short explanation in the text (lines 314-5).
Minor comments:
The authors should describe the nature of the NASP alleles in the main text and present evidence of robust NASP depletion, potentially both in ovaries and in embryos. The antibody works well for westerns (Fig S2B). This is sort of demonstrated later in Figure 4A, but only in NAAP x twine activated eggs.
Lines 163, 251, 339: minor typos Line 184: It would help to clarify- I'm assuming cytoplasmic concentration (or overall) rather than nuclear concentration. If nuclear, I'd expect the opposite relationship. This occurs again when discussing NASP (line 267). I suspect it's also not absolute concentration, but relative concentration difference between cytoplasm and nucleus. It would help clarify if the authors were more precise. Line 189: Given that the "established integrative model" helps to reject the hypothesis that NASP is involved in H3 import, I think it's important to describe the model a little more, even though it's previously published. Line 203: "The measured rate of H3.2 export from the nucleus is negligible" clarify this is in WT situations and not a conclusion from this study. Line 201: How can the authors be so sure that the decrease in WT is due to "the loss of non-chromatin bound nucleoplasmid H3.2-Dendra2?" Line 217: In the conclusion, the authors indicate that NASP indirectly affects soluble supply of H3 in the nucleoplasm. I do believe they've shown that the import rate effect is indirect, but I don't know why they conclude that the effect of NASP on the soluble nucleoplasmic H3 supply is indirect. Similarly, the conclusion is indirect on line 239. Yet, the authors have not shown it's not direct, just assumed since NASP results in 50% decrease to deposited maternal histones. Line 292: What is the nature of the NASP "mutant?" Is it a null? Similarly, what kind of "mutant" is the twine allele? Line 295. Line 316: Why did the authors use stage 14 egg chambers here when they previously used embryos? This becomes more clear later shortly, when the authors examine activated eggs, but it's confusing in text. Lines 343-348: It's unclear if the authors are drawing extended conclusions here or if they are drawing from prior literature (if so, citations would be required). For example, why during oogenesis/embryogenesis are aggregation and degradation developmentally separated? Lines 386-7: I do not understand why the authors conclude that H3 aggregation and degradation are "developmentally uncoupled" and why, in the absence of NASP, "H3 aggregation precedes degradation." Line 395: Why suddenly propose that NASP also functions in the nucleus to prevent aggregation, when earlier the authors suggest it functions only in the cytoplasm? Lines 409-413: The authors claim that histone deficiency likely does not cause the embryonic arrest seen in embryos from NASP mutant mothers. This is because H3 is reduced by 50% yet some embryos arrest long before they've depleted this supply. However, the authors also showed that H3 import rates are affected in these embryos due to lower H3 concentration. Since the early embryo cycles are so rapid, reduced H3 import rates could lead to early arrest, even though available H3 remains in the cytoplasm.
Significance
The significance of the work is conceptual, as NASP is known to function in H3 availability but the precise mechanism is elusive. This work represents a necessary advance, especially to show that NASP does not affect H3 import rates, nor does it chaperone H3 into the nucleus. However, the authors acknowledge that many questions remain. Foremost, why is NASP imported into the nucleus and what is its role there?
I believe this work will be of interest to those who focus on early animal development, but NASP may also represent a tool, as the authors conclude in their discussion, to reduce histone levels during development and examine nucleosome positioning. This may be of interest to those who work on chromatin accessibility and zygotic genome activation.
I am a genetics expert who works in Drosophila embryogenesis. I do not have the expertise to evaluate the aggregate methods presented in Figure 4.
-
-
publish.obsidian.md publish.obsidian.md
-
The repression of the Albigensians or Cathars ("pure ones") in southern France was an internal Christian crusade aimed at ridding the faith of people who were understood not only as heretics, but as agents of Satan who could damn entire communities.
It’s interesting that this crusade was Christians attacking other Christians just because they believed differently.
-
-
www.biorxiv.org www.biorxiv.org
-
Reviewer #1 (Public review):
Summary:
This paper presents an ambitious and technically impressive attempt to map how well humans can discriminate between colours across the entire isoluminant plane. The authors introduce a novel Wishart Process Psychophysical Model (WPPM) - a Bayesian method that estimates how visual noise varies across colour space. Using an adaptive sampling procedure, they then obtain a dense set of discrimination thresholds from relatively few trials, producing a smooth, continuous map of perceptual sensitivity. They validate their procedure by comparing actual and predicted thresholds at an independent set of sample points. The work is a valuable contribution to computational psychophysics and offers a promising framework for modelling other perceptual stimulus fields more generally.
Strengths:
The approach is elegant and well-described (I learned a lot!), and the data are of high quality. The writing throughout is clear, and the figures are clean (elegant in fact) and do a good job of explaining how the analysis was performed. The whole paper is tremendously thorough, and the technical appendices and attention to detail are impressive (for example, a huge amount of data about calibration, variability of the stim system over time, etc). This should be a touchstone for other papers that use calibrated colour stimuli.
Weaknesses:
Overall, the paper works as a general validation of the WPPM approach. Importantly, the authors validate the model for the particular stimuli that they use by testing model predictions against novel sample locations that were not part of the fitting procedure (Figure 2). The agreement is pretty good, and there is no overall bias (perhaps local bias?), but they do note a statistically-significant deviation in the shape of the threshold ellipses. The data also deviate significantly from historical measurements, and I think the paper would be considerably stronger with additional analyses to test the generality of its conclusions and to make clearer how they connect with classical colour vision research. In particular, three points could use some extra work:
(1) Smoothness prior.<br /> The WPPM assumes that perceptual noise changes smoothly across colour space, but the degree of smoothness (the eta parameter) must affect the results. I did not see an analysis of its effects - it seems to be fixed at 0.5 (line 650). The authors claim that because the confidence intervals of the MOCS and the model thresholds overlap (line 223), the smoothing is not a problem, but this might just be because the thresholds are noisy. A systematic analysis varying this parameter (or at least testing a few other values), and reporting both predictive accuracy and anisotropy magnitude, would clarify whether the model's smoothness assumption is permitting or suppressing genuine structure in the data. Is the gamma parameter also similarly important? In particular, does changing the underlying smoothness constraint alter the systematic deviation between the model and the MOCS thresholds? The authors have thought about this (of course! - line 224), but also note a discrepancy (line 238). I also wonder if it would be possible to do some analysis on the posterior, which might also show if there are some regions of color space where this matters more than others? The reason for doing this is, in part, motivated by the third point below - it's not clear how well the fits here agree with historical data.
(2) Comparison with simpler models. It would help to see whether the full WPPM is genuinely required. Clearly, the data (both here and from historical papers) require some sort of anisotropy in the fitting - the sensitivities decrease as the stimuli move away from the adaptation point. But it's >not< clear how much the fits benefit from the full parameterisation used here. Perhaps fits for a small hierarchy of simpler models - starting with isotropic Gaussian noise (as a sort of 'null baseline') and progressing to a few low-dimensional variants - would reveal how much predictive power is gained by adding spatially varying anisotropy. This would demonstrate that the model's complexity is justified by the data.
(3) Quantitative comparison to historical data. The paper currently compares its results to MacAdam, Krauskopf & Karl, and Danilova & Mollon only by visual inspection. It is hard to extract and scale actual data from historical papers, but from the quality of the plotting here, it looks like the authors have achieved this, and so quantitative comparisons are possible. The MacAdam data comparisons are pretty interesting - in particular, the orientations of the long axes of the threshold ellipses do not really seem to line up between the two datasets - and I thought that the orientation of those ellipses was a critical feature of the MacAdam data. Quantitative comparisons (perhaps overall correlations, which should be immune to scaling issues, axis-ratio, orientation, or RMS differences) would give concrete measures of the quality of the model. I know the authors spend a lot of time comparing to the CIE data, and this is great.... But re-expressing the fitted thresholds in CIE or DKL coordinates, and comparing them directly with classical datasets, would make the paper's claims of "agreement" much more convincing.
Overall, this is a creative and technically sophisticated paper that will be of broad interest to vision scientists. It is probably already a definitive methods paper showing how we can sample sensitivity accurately across colour space (and other visual stimulus spaces). But I think that until the comparison with historical datasets is made clear (and, for example, how the optimal smoothness parameters are estimated), it has slightly less to tell us about human colour vision. This might actually be fine - perhaps we just need the methods?
Related to this, I'd also note that the authors chose a very non-standard stimulus to perform these measurements with (a rendered 3D 'Greebley' blob). This does have the advantage of some sort of ecological validity. But it has the significant >disadvantage< that it is unlike all the other (much simpler) stimuli that have been used in the past - and this is likely to be one of the reasons why the current (fitted) data do not seem to sit in very good agreement with historical measurements.
-
-
www.biorxiv.org www.biorxiv.org
-
Reviewer #1 (Public review):
In this paper, the authors wished to determine human visuomotor mismatch responses in EEG in a VR setting. Participants were required to walk around a virtual corridor, where a mismatch was created by halting the display for 0.5s. This occurred every 10-15 seconds. They observe an occipital mismatch signal at 180 ms. They determine the specificity of this signal to visuomotor mismatch by subsequently playing back the same recording passively. They also show qualitatively that the mismatch response is larger than one generated in a standard auditory oddball paradigm. They conclude that humans therefore exhibit visuomotor mismatch responses like mice, and that this may provide an especially powerful paradigm for studying prediction error more generally.
Asking about the role of visuomotor prediction in sensory processing is of fundamental importance to understanding perception and action control, but I wasn't entirely sure what to conclude from the present paradigm or findings. Visuomotor prediction did not appear to have been functionally isolated. I hope the comments below are helpful.
(1) First, isolating visuomotor prediction by contrasting against a condition where the same video stream is played back subsequently does not seem to isolate visuomotor prediction. This condition always comes second, and therefore, predictability (rather than specifically visuomotor predictability) differs. Participants can learn to expect these screen freezes every 10-15 s, even precisely where they are in the session, and this will reduce the prediction error across time. Therefore, the smaller response in the passive condition may be partly explained by such learning. It's impossible to fully remove this confound, because the authors currently play back the visual specifics from the visuomotor condition, but given that the visuomotor correspondences are otherwise pretty stable, they could have an additional control condition where someone else's visual trace is played back instead of their own, and order counterbalanced. Learning that the freezes occur every 10-15 s, or even precisely where they occur, therefore, could not explain condition differences. At a minimum, it would be nice to see the traces for the first and second half of each session to see the extent to which the mismatch response gets smaller. This won't control for learning about the specific separations of the freezes, but it's a step up from the current information.
(2) Second, the authors admirably modified their visual-only condition to remove nausea from 6 df of movement (3D position, pitch, yaw, and roll). However, despite the fact it's far from ideal to have nauseous participants, it would appear from the figures that these modifications may have changed the responses (despite some pairwise lack of significance with small N). Specifically, the trace in S3 (6DOF) and 2E look similar - i.e., comparing the visuomotor condition to the visual condition that matches. Mismatch at 4/5 microvolts in both. Do these significantly differ from each other?
(3) It generally seems that if the authors wish to suggest that this paradigm can be used to study prediction error responses, they need to have controlled for the actions performed and the visual events. This logic is outlined in Press, Thomas, and Yon (2023), Neurosci Biobehav Rev, and Press, Kok, and Yon (2020) Trends Cogn Sci ('learning to perceive and perceiving to learn'). For example, always requiring Ps to walk and always concurrently playing similar visual events, but modifying the extent to which the visual events can be anticipated based on action. Otherwise, it seems more accurately described as a paradigm to study the influence of action on perception, which will be generated by a number of intertwined underlying mechanisms.
More minor points:
(1) I was also wondering whether the authors may consider the findings in frontal electrodes more closely. Within the statistical tests of the frontal electrodes against 0, as displayed in Figure 3c, the insignificance of the effect of Fp2 seems attributable to the small included sample size of just 13 participants for this electrode, as listed in Table S1, in combination with a single outlier skewing the result. The small sample size stands out especially in comparison to the sample size at occipital electrodes, which is double and therefore enjoys far more statistical power. It looks like the selected time window is not perfectly aligned for determining a frontal effect, and also the distribution in 3B looks like responses are absent in more central electrodes but present in occipital and frontal ones. I realise the focus of analysis is on visual processing, but there are likely to be researchers who find the frontal effect just as interesting.
(2) It is claimed throughout the manuscript that the 'strongest predictor (of sensory input) - by consistency of coupling - is self-generated movement'. This claim is going to be hard to validate, and I wonder whether it might be received better by the community to be framed as an especially strong predictor rather than necessarily the strongest. If I hear an ambulance siren, this is an especially strong predictor of subsequent visual events. If I see a traffic light turn red, then yellow, I can be pretty certain what will happen next. Etc.
(3) The checkerboard inversion response at 48 ms is incredibly rapid. Can the authors comment more on what may drive this exceptionally fast response? It was my understanding that responses in this time window can only be isolated with human EEG by presenting spatially polarized events (cf. c1, e.g., Alilovic, Timmermans, Reteig, van Gaal, Slagter, 2019, Cerebral Cortex)
-
-
www.biorxiv.org www.biorxiv.org
-
Reviewer #1 (Public review):
Summary:
The authors show that the lower frequency (~5Hz) stimulation of the intermittent theta-burst stimulation (iTBS) via repetitive transcranial magnetic stimulation (rTMS) serves as a more effective stimulation paradigm than the high-frequency protocols (HF-rTMS, ~10Hz) with enhancing plasticity effects via long-term potentiation (LTP) and depression (LTD) mechanisms. They show that the 5 Hz patterned pulse structure of the iTBS is an exact subharmonic of the 10 Hz high-frequency rTMS, creating a connection between the two paradigms and acting upon the same underlying synchrony mechanism of the dominant alpha-rhythm of the corticothalamic circuit.
First, the authors create a corticothalamic neural population model consisting of 4 populations: cortical excitatory pyramidal and inhibitory interneuron, and thalamic excitatory relay and inhibitory reticular populations. Second, the authors include a calcium-dependent plasticity model, in which calcium-related NMDAR-dependent synaptic changes are implemented using a BCM metaplasticity rule. The rTMS-induced fluctuations in intracellular calcium concentrations determine the synaptic plasticity effects.
Strengths:
The model (corticothalamic neural population with calcium-dependent plasticity, with TBS input for rTMS) is thoroughly built and analyzed.
The conclusions seem sound and justified. The authors justifiably link stimulation parameters (especially the alpha subharmonics iTBS frequency) with fluctuations in calcium concentration and their effects on LTP and LTD in relevant parts of the corticothalamic circuit populations leading to a dampening of corticothalamic loop gains and enhancement of intrathalamic gains with an overall circuit-wide feedforward inhibition (= inhibitory activity is enhanced via excitatory inputs onto inhibitory neurons) and a resulting suppression of the activity power. In other words: alpha-resonant iTBS protocols achieve broadband power suppression via selective modulation of corticothalamic FFI.
(1) The model is well-described, with the model equations in the main text and the parameters in well-formatted tables.
(2) The relationship between iTBS timing and the phase of rhythms is well explained conceptually.
(3) Metaplasticity and feedforward inhibition regulation as a driver for the efficacy of iTBS are well explored in the paper.
(4) Efficacy of TBS, being based on mimicry of endogenous theta patterns, seems well supported by this simulation.
(5) Recovery between periods of calcium influx as an explanation for why intermittency produces LTP effects where continuous stimulation fails is a good justification for calcium-based metaplasticity, as well as for the role of specific pulse rate.
(6) Circuit resonance conclusion is interesting as a modulating factor; the paper supports this hypothesis well.
(7) The analysis of corticothalamic dampening and intrathalamic enhancement in the 3D XYZ loop gain space is a strong aspect of the paper.
Weaknesses:
(1) Overall, the paper is difficult to follow narratively - the motivation (formulated as a specific research question) for each section can be a bit unclear. The paper could benefit from a minor rewrite at the start of each section to justify each section's reasoning. The Discussion is too long and should be shortened and limited to the main points.
(2) While the paper refers to modelling and data in discussion, there is no direct comparison of the simulations in the figures to data or other models, so it's difficult to evaluate directly how well the modelling fits either the existing model space or data from this region. Where exactly the model/plasticity parameters from Table 5 and the NFTsim library come from is not easy to find. The authors should make the link from those parameters to experimental data clearer. For example, which clinical or experimental data are their simulations of the resting-state broadband power suppression based on?
(3) The figures should be modified to make them more understandable and readable.
(4) The claim in the abstract that the paper introduces "a novel paradigm for individualizing iTBS treatments" is too strong and sounds like overselling. The paper is not the first computational modelling of TBS - as acknowledged also by the authors when citing previous mean-field plasiticity modelling articles. Btw. the authors could briefly mention and include also references also to biophysically more detailed multi-scale approaches such as https://doi.org/10.1016/j.brs.2021.09.004 and https://doi.org/10.1101/2024.07.03.601851 and https://doi.org/10.1016/j.brs.2018.03.010
(5) The modelling assumes the same CaDP model/mechanism for all excitatory synapses/afferents. How well is this supported by experimental evidence? Have all excitatory synaptic connections in the cortico-thalamic circuit been shown to express CaDP and metaplasticity? If not, these limitations (or predictions of the model) should be mentioned. Why were LTP calcium volumes never induced within thalamic relay-afferent connections se and sr? What about inhibitory synapses in the circuit model? Were they plastic or fixed?
(6) Minor point: Metaplasticity is modelled as an activity-dependent shift in NMDAR conductance, which is supported by some evidence, but there are other metaplasticity mechanisms. Altering NMDA-synapse affects also directly synaptic AMPA/NMDA weight and ratio (which has not been modelled in the paper). Would the model still work using other - more phenomenological implementation of the sliding threshold - e.g. based on shifting calcium-dependent LTP/LTD windows or thresholds (for a phenomenological model of spike/voltage-based STDP-BCM rules, see https://doi.org/10.1007/s10827-006-0002-x and https://doi.org/10.1371/journal.pcbi.1004588) - maybe using a metaplasticity extension of Graupner and Brunel CaDP model. A brief discussion of these issues might be added to the manuscript - but this is just a suggestion.
(7) Short-term plasticity (depression/facilitation) of synapses is neglected in the model. This limitation should be mentioned because adding short-term synaptic dynamics might affect strongly circuite model dynamics.
-
-
go.gale.com go.gale.com
-
I focus on the essay of only one student, Robert (pseudonym), because his analysis of a hip-hop text led me to the serious study of scholarship on hip-hop language and literacy
I think this is a really powerful sentence, it's not just saying "oh I thought this student wrote a good paper" it's saying that it was more then just a paper is was a catalyst for the author to change paths, it wasn't just inspiring it was moving enough to research and make a career out of it.
-
-
www.jstor.org www.jstor.org
-
I feel like if I can't write it's a lot of things inside me, a lot offeelings that I can't, um, I can't send them to other people, you know. 'Cause, uh,I find myself when I'm writing.
writing portrayed as more than just a leisure activity or hobby. It is a way of finding the hidden parts of oneself.
-
-
allpoetry.com allpoetry.com
-
This sleep of mine, whatever sleep it is. Were he not gone, The woodchuck could say whether it's like his Long sleep, as I describe its coming on,
These lines are incredibly romantic because Frost wonders what kind of sleep he's falling into and whether its more than just "sleep". He doesn't know if it'll be normal or death. He also compares himself to a woodchuck which are animals that hibernate just like the "long sleep" Frost describes which is almost like them "dying" and coming back when it's spring.
-
-
social-media-ethics-automation.github.io social-media-ethics-automation.github.io
-
Reintegration “Public shaming must aim at, and make possible, the reintegration of the norm violator back into the community, rather than permanently stigmatizing them.”
I think it’s ironic that public shaming is supposed to leave room for people to eventually rejoin the community, because social media basically gives them no space to do that. The reading says shaming should allow for the possibility of reintegration, but online most people just want entertainment and don’t care whether someone ever gets the chance to repair anything. It makes the internet feel like a place that amplifies the “shame” part while intentionally deleting the “repair” part.
-
-
pubs.lib.uiowa.edu pubs.lib.uiowa.edu
-
(3) ChrisAnn Cleland, a real estate agent from Virginia, express disappointment aboutPresident Obama’s economic plan in an interview with the Washington Post (Rich):“Nothing’s changed for the common guy,” she said. “I feel likeI’ve been punked.”(4) Referencing Cleland’s remark, the title of New York Times columnist FrankRich’s Op-ed article asks: “Is Obama Punking Us?” Rich writes in the last para-graph of his article:“The larger fear is that Obama might be just another corporatist,punking voters much as the Republicans do when they claim tobe all for the common guy.”
Example – Word “punked,” which comes from Black language, being used by white professionals in mainstream media. Shows how Black English moves into “respectable” spaces while students are still told it’s not appropriate for school writing.
-
-
www.biorxiv.org www.biorxiv.org
-
Note: This response was posted by the corresponding author to Review Commons. The content has not been altered except for formatting.
Learn more at Review Commons
Reply to the reviewers
Review report for 'Sterols regulate ciliary membrane dynamics and hedgehog signaling in health and disease', Lamazière et al.
Reviewer #1
In this manuscript, Lamazière et al. address an important understudied aspect of primary cilium biology, namely the sterol composition in the ciliary membrane. It is known that sterols especially play an important role in signal transduction between PTCH1 and SMO, two upstream components of the Hedgehog pathway, at the primary cilium. Moreover, several syndromes linked to cholesterol biosynthesis defects present clinical phenotypes indicative of altered Hh signal transduction. To understand the link between ciliary membrane sterol composition and Hh signal transduction in health and disease, the authors developed a method to isolate primary cilia from MDCK cells and coupled this to quantitative metabolomics. The results were validated using biophysical methods and cellular Hh signaling assays. While this is an interesting study, it is not clear from the presented data how general the findings are: can cilia be isolated from different mammalian cell types using this protocol? Is the sterol composition of MDCK cells expected to the be the same in fibroblasts or other cell types? Without this information, it is difficult to judge whether the conclusions reached in fibroblasts are indeed directly related to the sterol composition detected in MDCK cells. Below is a detailed breakdown of suggested textual changes and experimental validations to strengthen the conclusions of the manuscript.
We would like to thank the reviewer for their helpful comments
Major comments:
- It appears that the comparison has been made between ciliary membranes and the rest of the cell's membranes, which includes many other membranes besides the plasma membrane. This significantly weakens the conclusions on the sterol content specific to the cilium, as it may in fact be highly similar to the rest of the plasma membrane. It is for example known that lathosterol is biosynthesized in the ER, and therefore the non-presence in the cilium may reflect a high abundance in the ER but not necessarily in the plasma membrane.
The reviewer is correct that we compared the sterol composition of the primary ciliary membrane to the average of the remaining cellular membranes. We agree that this broader reference fraction contains multiple intracellular membranes, including ER- and Golgi-derived compartments, and therefore does not isolate the plasma membrane specifically. We would like to emphasize that our study did not aim to compare the cilium directly to the plasma membrane, nor did we claim that the comparison was in any way related to the plasma membrane. It is also worth noting that previous studies in other ciliated organisms have reported a higher cholesterol content in cilia compared to the plasma membrane, suggesting that the two membranes may not be compositionally identical despite their continuity. However, we concur that determining the sterol composition of the MDCK plasma membrane would provide valuable context and enable a comparison with the membrane continuous with the ciliary membrane. Hence, we are willing to try isolating plasma membrane in the same cellular contexts.
- While the protocol to isolate primary cilium from MDCK cells is a valuable addition to the methods available, it would be good to at least include a discussion on its general applicability. Have the authors tried to use this protocol on fibroblasts for example?
Thank you for the reviewer's positive comment on the value of the ciliary isolation protocol. Indeed, we have attempted to apply the same approach to other ciliated cell types, namely IMCD3 and MEF cells. In the case of IMCD3 cells, we were able to isolate primary cilia using the same general strategy; however, we are still refining the preparation, as the overall yield is lower than in MDCK cells and the amount of material obtained is currently insufficient for comprehensive biochemical analyses. With MEF (fibroblast) cells, the procedure proved even more challenging, as the yield of isolated cilia was extremely low. This difficulty is likely due to the shorter length of fibroblast cilia and to their positioning beneath the cell body, which probably makes them more resistant to detachment. Overall, these observations suggest that while the protocol can be adapted to other cell types, its efficiency depends on cellular architecture. We have added a discussion of these aspects in the revised manuscript to clarify the method's current scope and limitations (lines 492-502).
- Some of the conclusions in the introduction (lines 75-80) seem to be incorrectly phrased based on the data: in basal conditions, ciliary membranes are already enriched in cholesterol and desmosterol, and the treatment lowers this in all membranes.
We agree, this was modified in the revised manuscript (lines 75-80).
- There seems to be little effect of simvastatin on overall cholesterol levels. Can the authors comment on this result? How would the membrane fluidity be altered when mimicking simvastatin-induced composition? Since the effect on Hh signaling appears to be the biggest (Figure 5B) under simvastatin treatment, it would be interesting to compare this against that found for AY9944 treatment. Also, the authors conclude that the effects of simvastatin treatment on ciliary membrane sterol composition are the mildest, however, one could argue that they are the strongest as there is a complete lack of desmosterol.
We thank the reviewer for these insightful comments. Regarding the modest overall effect of simvastatin on cholesterol levels, we would like to note that MDCK cells are an immortalized epithelial cell line with high metabolic plasticity. Such cancer-like cell types are known to exhibit enhanced de novo lipogenesis, particularly under culture conditions with ample glucose availability. This compensatory lipid biosynthesis can partially counterbalance pharmacological inhibition of the cholesterol biosynthetic pathway. Because simvastatin acts upstream in the pathway (at HMG-CoA reductase), its inhibition primarily reduces early intermediates rather than fully depleting end-product cholesterol, explaining the relatively mild changes observed in total cholesterol content.
Concerning desmosterol, we agree with the reviewer that its complete loss under simvastatin treatment is a striking finding that deserves further discussion. Interestingly, our data show that simvastatin treatment produces the strongest inhibition of pathway activation (as measured by SMO activation), but the weakest effect on signal transduction downstream of constitutively active SMOM2. This dichotomy suggests that the absence of desmosterol may preferentially affect the activation step of Hedgehog signaling at the ciliary membrane, without equally impacting downstream propagation. We have expanded the Result section to highlight this potential role of desmosterol in the activation phase of Hedgehog signaling and to contrast it with the effects observed under AY9944 treatment (lines 463-469).
It is not clear to me why the authors have chosen to use SAG to activate the Hh pathway, as this is a downstream mode of activation and bypasses PTCH1 (and therefore a potentially sterol-mediated interaction between the two proteins). It would be very informative to compare the effect of sterol modulation on the ability of ShhN vs SAG to activate the pathway.
Our study aims to demonstrate that the sterol composition of the ciliary membrane plays an essential role in the proper functioning of the Hedgehog (Hh) signaling pathway, comparable in importance to that of oxysterols and free cholesterol. Because ShhN itself is covalently modified by cholesterol, and Smoothened (SMO) can be directly activated by both oxysterols and cholesterol, we reasoned that using a non-native SMO agonist such as SAG would allow us to specifically assess defects arising from alterations in membrane-bound sterols. In this way, pathway activation by SAG provides a more direct readout of the functional contribution of ciliary membrane sterols to SMO activity, independent of potential confounding effects related to ShhN processing, secretion, or PTCH1-mediated regulation.
- The conclusions about the effect of tamoxifen on SMO trafficking in MEFs should be validated in human patient cells before being able to conclude that there is a potential off-target effect (line 438). Also, if that is the case, the experiment of tamoxifen treatment of EBP KO cells should give an additional effect on SMO trafficking. Also, could the CDPX2 phenotypes in patients be the result of different cell types being affected than the fibroblast used in this study?
We agree that carrying the proposed experiment would be a good way to assess a potential off-target effect. However, such validation is beyond the scope of the present study, as this comment on off-target effect was aimed primarily to propose a mechanistic hypothesis to explain the differences observed in Hedgehog pathway activation between patient-derived fibroblasts and tamoxifen-treated MEFs. We leaned towards this hypothesis because drug treatments are known for their overall variable specificity, but we agree other hypotheses are possible, and among them the difference in cell type, as both are fibroblasts but from different origin. We rephrased this passage in the revised manuscript (lines 447-448 ).
Regarding the reviewer's third point, we fully agree that the CDPX2 phenotype in patients is unlikely to arise solely from fibroblast dysfunction. Nevertheless, fibroblasts are the only patient-derived cells currently available to us, and they provide a useful model for assessing ciliary signaling. It is reasonable to expect that similar defects could occur in other, more physiologically relevant cell types.
- For the experiments with the SMO-M2 mutant, it would be useful to show the extent of pathway activation by the mutant compared to SAG or ShhN treatment of non-transfected cells. Moreover, it will be necessary to exclude any direct effects of the compound treatment on the ability of this mutant to traffic to the primary cilium, which can easily be done using fluorescence microscopy as the mutant is tagged with mCherry.
The SmoM2 mutant is indeed a well-characterized constitutively active form of Smoothened that has been extensively studied by us and others. It is well established that this mutant correctly localizes to the primary cilium and robustly activates the Hedgehog pathway in MEFs (see Eguether et al., Dev. Cell, 2014 or Eguether et al, mol.biol.cell, 2018). In our study, we have already included supporting evidence for pathway activation in Supplementary Figure S1b, showing Gli1 expression levels in untreated MEFs transfected with SmoM2, which illustrates the extent of its activation compared to ligand-induced conditions.
In line with the reviewer's recommendation, we will additionally include microscopy data showing SmoM2 localization in MEFs treated with the different sterol modulators. These data should confirm that the observed effects are not due to altered ciliary trafficking of the mutant protein but instead reflect changes in downstream signaling or membrane composition.
Minor comments:
Line 74: 'in patients', should be rephrased to 'patient-derived cells'
This was modified in the revised manuscript
Figure 2A: What do the '+/-' indicate? They seem to be erroneously placed.
We apologize for the oversight, the figures initially submitted with the manuscript inadvertently included some earlier versions, which explains several of the discrepancies noted by the reviewers. This issue has been corrected in the revised submission, and all figures have now been updated to reflect the finalized data.
Figure 2B: no label present for which bar represents cilia/other membranes
We apologize for the oversight, the figures initially submitted with the manuscript inadvertently included some earlier versions, which explains several of the discrepancies noted by the reviewers. This issue has been corrected in the revised submission, and all figures have now been updated to reflect the finalized data.
Figure 2C: this representation is slightly deceptive, since the difference between cells and cilia for lanosterol is not significantly different as shown in figure 2A.
This representation has been removed in the revised figures.
Figure 3A: it would be useful to also show where 8-DHC is in the biosynthetic pathway.
This has been modified in the revised figures.
Line 373: the title should be rephrased as it infers that DHCR7 was blocked in model membranes, which is not the case.
This has been modified in the revised manuscript.
Lines 377-384: this paragraph seems to be a mix of methods and some explanation, but should be rephrased for clarity.
We believe the technical information within this paragraph are useful for the understanding of the reader. We would rather leave as is unless recommended by other reviewers or editorial staff.
Line 403: 'which could explain the resulting defects in Hedgehog signaling': how and what defects? At this point in the study no defects in Hh signaling have been shown.
This has been modified in the revised manuscript.
Figure 4D: 'd' is missing
We apologize for the oversight, the figures initially submitted with the manuscript inadvertently included some earlier versions, which explains several of the discrepancies noted by the reviewers. This issue has been corrected in the revised submission, and all figures have now been updated to reflect the finalized data.
Line 408: SAG treatment resulted in slightly shorter cilia: this is not the case for just SAG treated cilia, but only for the combination of SAG + AY9944. However, in that condition there appears to be a subpopulation of very short cilia, are those real?
This is correct, this is not the case for untreated cilia, but the short population is real, not only in AY9944 but also in Tamoxifen and Simvastatin. Again, the relevance and significance of minor cilia length change is unclear and we are not trying to draw any other conclusion from this than saying that the ciliary compartment is modified.
Figure 5b: it would be good to add that all conditions contained SAG.
This has been modified in the revised figures.
Figure 5D: Since it is shown in Fig 5C that there are no positive cilia -SAG, there is no point to have empty graphs in Fig 5D on the left side, nor can any statistics be done. Similarly for 5K.
We think this is still worth having in the figure. As the reviewer noted in one of his next comment, there are cases where Smoothened or Patched can be abnormally distributed (see also Eguether et al, mol biol cell, 2018). This shows that we checked all conditions for presence or absence of Smo and that there is no signal to be found. We would rather leave it as is unless asked otherwise by editorial staff.
Figure 5E: it is not clearly indicated what is visualized in the inserts, sometimes it's a box, sometimes a line and they seem randomly integrated into the images.
We apologize for the oversight - the figures initially submitted with the manuscript inadvertently included some earlier versions, which explains several of the discrepancies noted by the reviewers. This issue has been corrected in the revised submission, and all figures have now been updated to reflect the finalized data.
Figure 5H: is this the intensity in just SMO positive cilia? If yes, this should be indicated, and the line at '0' for WT-SAG should be removed. I am also surprised there is then ns found for WT vs SLO, since in WT there are no positive cilia, but in SLO there are a few, so it appears to be more of a black-white situation. Perhaps it would be useful to split the data from different experiments to see if it consistently the case that there is a low percentage of SMO positive cilia in SLO cells.
Yes, as in the rest of figure 5, the fluorescence intensity of Smo is only taken into account in SMO positive cells. This is now indicated in figure legend (lines 890, 898, 903 ). As for Smo positive, this is a good suggestion. We checked and for cilia in non-activated SLO patients, there are 8 positive cilia over a total of 240 counted cilia, mainly from one of the experiments. We could remove the data or leave as is given that the result is not significant.
Fig S1: panels are inverted compared to mentioning in the text.
We apologize for the oversight, the figures initially submitted with the manuscript inadvertently included some earlier versions, which explains several of the discrepancies noted by the reviewers. This issue has been corrected in the revised submission, and all figures have now been updated to reflect the finalized data.
Methods-pharmacological treatments: there appear to be large differences in concentrations chosen to treat MDCK versus MEF cells - can the authors comment on these choices and show that the enzymes are indeed inhibited at the indicated concentrations?
We thank the reviewer for this important comment. The concentrations of the pharmacological treatments were optimized separately for MDCK and MEF cells based on cell-type-specific tolerance. For each compound, we used the highest concentration that produced no detectable cytotoxicity or morphological changes. These conditions ensured that the treatments were effective (as seen by changes in sterol composition in MDCK cilia and Hh pathway phenotypes in treated MEFs) and compatible with cell viability and ciliation. Although we did not directly assay enzymatic inhibition in each case, the selected concentrations are consistent with those previously reported to inhibit the targeted enzymes in similar cellular contexts.
Compound
Typical Concentration Range in Mammalian Cell Culture
Typical Exposure Duration
Example Cell Types
Representative Peer-Reviewed References
AY9944 (DHCR7 inhibitor)
1-10 µM widely used; 1 µM for minimal on-target effects; 2.5-10 µM for robust sterol shifts
24-72 h; some sterol studies up to several days
HEK293, fibroblasts, neuronal cells, macrophages
Kim et al., J Biol Chem, 2001 - used 1 µM in dose-response experiments.; Haas et al., Hum Mol Genet, 2007 - 1 µM in cell-based assays.; Recent macrophage sterol study - 2.5-10 µM to induce 7-DHC accumulation.
Simvastatin (HMG-CoA reductase inhibitor)
0.1-10 µM common; 1-10 µM most widely used for robust pathway inhibition
24-72 h
Diverse mammalian lines, including liver, fibroblasts, epithelial cells
Bytautaite et al., Cells (2020) - discusses common in-vitro ranges (1-10 µM).; Mullen et al., 2011 - used 10 µM simvastatin, noting it is a standard in-vitro concentration.
Tamoxifen (modulator of sterol metabolism)
1-20 µM; 1-5 µM for mild/longer treatments; 10-20 µM in cancer/cilia signaling studies
24-72 h (longer treatments often at 1-5 µM)
MDCK, MEFs, MCF-7, diverse epithelial lines
Schlottmann et al., Cells (2022) - used 5-25 µM in sterol-related cell studies.; MCF-7 literature - 0.1-1 µM for estrogenic signaling, higher (5-10 µM) for metabolic/sterol pathway effects.; Additional cancer cell work indicating similar ranges.
This information has been clarified in the revised Methods section (lines 222-224).
(optional): it would be interesting to include a gamma-tubulin staining on the cilium prep to see if there is indeed a presence of the basal body as suggested by the proteomics data.
Thank you, we will try this.
There are many spelling mistakes and inconsistencies throughout the manuscript and its figures (mix of French and English for example) so careful proofreading would be warranted. Moreover, there are many mentionings of 'Hedgehog defects' or 'Hedgehog-linked', where in fact it is a defect in or link to the Hedgehog pathway, not the protein itself. This should be corrected.
We thank the reviewer for noting these issues. We apologize for the inconsistencies observed in the initial submission, as mentioned previously, some of the figures inadvertently included earlier versions, which may have contributed to the errors identified. All figures have now been carefully revised and updated in the resubmitted manuscript.
Regarding the text, we are surprised to hear about the spelling inconsistencies, as the manuscript was professionally proofread prior to submission (documentation can be provided upon request). Nevertheless, we have conducted an additional round of thorough proofreading to ensure consistency throughout the text and figures.
Finally, we have corrected all instances of "Hedgehog defects" or "Hedgehog-linked" to the more accurate phrasing "Hedgehog pathway defect" or "Hedgehog pathway-linked," as suggested by the reviewer throughout the manuscript.
Reviewer #1 (Significance (Required)):
The study of ciliary membrane composition is highly relevant to understand signal transduction in health and disease. As such, the topic of this manuscript is significant and timely. However, as indicated above, there are limitations to this study, most notably the comparison of ciliary membrane versus all cellular membranes (rather than the plasma membrane), which weakens the conclusions that can be drawn. Moreover, cell-type dependency should be more thoroughly addressed. There certainly is a methodological advance in the form of cilia isolation from MDCK cells, however, it is unclear how broadly applicable this is to other mammalian cell types.
We would like to thank the reviewer for their helpful comments and we appreciate the reviewer's recognition of the relevance and timeliness of studying ciliary membrane composition in the context of signaling regulation. We fully acknowledge that our comparison was made between the primary ciliary membrane and the total cellular membrane fraction, which encompasses multiple intracellular membranes. Our intent, however, was to obtain a global overview of how the ciliary membrane differs from the average membrane environment within the cell, thereby highlighting features that are unique to the cilium as a signaling organelle. This approach provides valuable baseline information that complements, rather than replaces, future targeted comparisons with the plasma membrane. As mentioned in this reply, we aim at carrying out these experiments before publication. Regarding cell-type dependency, we concur that ciliary lipid composition may vary between cell types, reflecting differences in their functional specialization. Our method was intentionally established in MDCK cells, which are epithelial and highly ciliated, to ensure sufficient yield and reproducibility. We have initiated trials with other mammalian cell types, including IMCD3 and MEF cells, and while yields remain limited, preliminary results indicate that the approach is adaptable with further optimization. Thus, our current work establishes a robust and reproducible proof of concept in a mammalian model, providing the first detailed sterol fingerprint of a mammalian primary cilium.
We believe this constitutes a significant methodological and conceptual advance, as it opens the way for systematic exploration of ciliary lipid composition across diverse mammalian systems and pathological contexts.
Reviewer #2 (Evidence, reproducibility and clarity (Required)):
Overview Accumulating evidence suggests that sterols play critical roles in signal transduction within the primary cilium, perhaps most notably in the Hedgehog cascade. However, the precise sterol composition of the primary cilium, and how it may change under distinct biological conditions, remains unknown, in part because of the lack of reproducible, widely accepted procedures to purify primary cilia from mammalian cultured cells. In the present study, the authors have designed a method to isolate the cilium from the MDCK cells efficiently and then utilized this procedure in conjunction with mass spectrometry to systematically analyze the sterol composition of the ciliary membrane, which they then compare to the sterol composition of the cell body. By analyzing this sterol profiling. the authors claim that the cilium has a distinct sterol composition from the cell body, including higher levels of cholesterol and desmosterol but lower levels of 8-DHC and & Lathosterol. This manuscript further demonstrates that alteration of sterol composition within cilia modulates Hedgehog signaling. These results strengthen the link between dysregulated Hedgehog signaling and defects in cholesterol biosynthesis pathways, as observed in SLOS and CDPX2.
While the ability to isolate primary cilia from cultured MDCK cells represents an important technical achievement, the central claim of the manuscript - that cilia have a different sterol composition from the cell body - is not adequately supported by the data, and more rigorous comparisons between the ciliary membrane and key organellar membranes (such as plasma membrane) are required to make this claim. Moreover, although the authors have repeatedly mention that the ciliary sterol composition is "tightly regulated" there is no evidence provided to support such claim. At best, the data suggest that the cilium and cell body may differ in sterol composition (though even that remains uncertain), but no underlying regulatory mechanisms are demonstrated. In addition, much of the 2nd half of the paper represents a rehash of experiments with sterol biosynthesis inhibitors that have already been published in the literature, making the conceptual advance modest at best. Lastly, the link between CDPX2 and defective Hedgehog signaling is tenuous.
We would like to thank the reviewer for their helpful comments
Major comments
Figure 1. C) Although the isolation of cilium from the MDCK cells using dibucaine treatment seems to be very efficient, the quality control of their fractionation procedure to monitor the isolation is limited to a single western blot of the purified cilia vs. cell body samples, with no representative data shown from the sucrose gradient fractionation steps. Given that prior studies (including those from the Marshall lab cited in this manuscript) found that 1) sucrose gradient fractionation was essential to obtain relatively pure ciliary fractions, and 2) the ciliary fractions appear to spread over many sucrose concentrations in those prior studies , the authors should have included the comparison of the fractionation profile from the sucrose gradient while isolating the primary cilium. This additional information would have further clarified and supported the efficiency of their proposed method.
We thank the reviewer for their insightful comments regarding the quality control of our ciliary fractionation. We would like to clarify several important methodological aspects that distinguish our approach from those used in the studies cited (including those from the Marshall lab). In the cited work, the authors used a continuous sucrose gradient ranging from 30 % to 45 %, which allowed visualization of the distribution of ciliary proteins across the gradient. In contrast, we employed a discontinuous sucrose gradient (25 % / 50 %) optimized for higher recovery and reproducibility in our hands. In our preparation, the primary cilia consistently localize at the interface between the 25 % and 50 % layers. We systematically collect five 1 mL fractions from this interface and use fractions 1-3 for downstream analyses, as fractions 4-5 are typically already depleted of ciliary material. This targeted collection ensures good enrichment and low contamination, while avoiding unnecessary dilution of the limited ciliary sample. We also note that the prior studies the reviewer refers to were optimized for proteomic analyses, and therefore used actin as a marker of contamination from the cell body. In our case, the downstream application is lipidomic profiling, for which such protein-based contamination markers are not directly informative, since no reliable lipid marker exists to differentiate between organelle membranes. For this reason, we limited the protein-level validation to a semi-quantitative assessment of ciliary enrichment using ARL13B Western blotting, which robustly reports the presence and enrichment of ciliary membranes. Finally, to complement this targeted validation, we performed proteomic analysis followed by Gene Ontology (GO) Enrichment Analysis using the PANTHER database. This analysis evaluates the overrepresentation of proteins associated with ciliary structures and functions relative to the background frequency in the Canis lupus familiaris proteome. The resulting enrichment profile confirms that the isolated material is highly enriched in ciliary components and somewhat depleted of non-ciliary contaminants, thereby serving as an unbiased and global assessment of sample specificity and purity. We believe that, together, these methodological choices provide a rigorous and quantitative validation of our fractionation efficiency and support the robustness of the cilia isolation protocol used in this study.
-
D) The authors presented proteomic data for the peptides analyzed from the isolated cilia in the form of GO term analysis; however, they did not provide examples of different proteins enriched within their fractionation procedure, aside from Arl13b shown in the blot. Including a summary table with representative proteins identified in the isolated ciliary fraction, along with the relative abundance or percentage distribution of these proteins, would make the data more informative.
We thank the reviewer for this valuable suggestion. As mentioned in the manuscript, our proteomic dataset includes numerous hallmark components of the cilium, such as 18 IFT proteins, 4 BBS proteins, and several Hedgehog pathway components (including SuFu and Arl13b), as well as axonemal (Tubulin, Kinesin, Dynein) and centrosomal proteins (Centrin, CEPs, γ-Tubulin, and associated factors). This composition demonstrates that the isolated fraction is highly enriched in bona fide ciliary components while retaining a small proportion of basal body proteins, which is expected given their physical continuity. Importantly, our dataset shows a 70% overlap with the ciliary proteome published by Ishikawa et al. and a 41% overlap with the CysCilia consortium's list of potential ciliary proteins, which supports both the specificity and reliability of our isolation procedure. Regarding the suggestion to present relative protein abundances, we would like to clarify that defining "relative to what" is challenging in this context. The stoichiometry of ciliary proteins is largely unknown, and relative abundance normalized to total protein content can be misleading, as ciliary structural and signaling components differ greatly in copy number and membrane association. For this reason, we chose to highlight in the text proteins such as BBS and IFTs, which are known to be of low abundance within the cilium; their detection supports the depth and specificity of our proteomic coverage. In addition, we performed an unbiased Gene Ontology (GO) Enrichment Analysis using the PANTHER database, which provides a systematic and quantitative overview of the biological processes and cellular components overrepresented in our dataset relative to the canine proteome. This analysis with regard to purity wa already discussed in the submitted manuscript discussion. To further address the reviewer's comment, we will include as a supplemental table in the revised manuscript, a summary table listing representative ciliary proteins identified in our fraction, including those overlapping with the CysCilia (Gold ans potential lists), CiliaCarta and Ishikawa/Marshall proteomes. This addition should make the dataset more transparent and informative while preserving scientific rigor.
Figure 2.
The authors represented the comparison of sterol content within the cilia versus whole cell (as cell membranes). Since different organelles have a very diverse degree of cholesterol contents within them, for instance plasma membrane itself is around 50 mol% cholesterol levels while organelles like ER have barely any cholesterol. Thus, comparing these two samples and claiming a 2.5-fold increase in cholesterol levels is misleading. A more appropriate comparison would be between isolated primary cilia and isolated plasma membranes (procedures to isolate plasma membranes have been described previously, e.g., Naito et al., eLife 2019; Das et al, PNAS 2013. The absence of such controls makes it difficult to fully validate the reported magnitude of sterols enrichment in cilia relative to the cell surface.
As already discussed above for reviewer 1, we would like to emphasize that our study did not aim to compare the cilium directly to the plasma membrane, nor did we claim that the comparison was in any way related to the plasma membrane. Our intent, was to obtain a global overview of how the ciliary membrane differs from the average membrane environment within the cell, thereby highlighting features that are unique to the cilium as a signaling organelle. This approach provides valuable baseline information that complements, rather than replaces, future targeted comparisons with the plasma membrane. However, we concur that determining the sterol composition of the MDCK plasma membrane would provide valuable context and enable a comparison with the membrane continuous with the ciliary membrane. Hence, we are willing to try isolating plasma membrane in the same cellular contexts, and we thank the reviewer for the proposed literature.
Also, because dibucaine was used here to isolate MDCK cilia, a control experiment to exclude possible effects of the dibucaine treatment on sterol biosynthesis would be helpful.
Thank you for this comment, we will verify this point by quantifying by GC-MS the sterol content of whole MDCK cells with and without 15 minutes-dibucaine treatments.
Figure 3.
Tamoxifen is a potent drug for nuclear hormone receptor activity and thus can independently influence various cellular processes. As several experiments in the later sections of the manuscript rely on tamoxifen treatment of cells, it is important that the authors include appropriate controls for tamoxifen treatment, to confirm that the observed effects do not stem from effects on nuclear hormone receptor activity. This would ensure that the observed effects can be confidently attributed to the experimental manipulation rather than to the intrinsic effects of tamoxifen.
The reviewer is right, tamoxifen, like many drugs, has pleiotropic effects in different cell processes. Aware of this possible issue, we turned to a genetic model creating a CRISPR-CAS9 mediated knock down of EBP, the enzyme targeted by tamoxifen. We showed in figure 5 that the results between tamoxifen treated cells and CRIPSR EBP cells were in accordance with one another, showing that, for hedgehog signaling, the effect of tamoxifen recapitulates the effect of the enzyme KO.
Figure4. The authors present the results of spectroscopy studies to analyze generalized polarization (GP) of liposomes in vitro , but only processed data are shown, and the raw spectra are not provided. The authors need to present representative spectra to enable the readers to interact the raw data from the experiments.
This has been added to new supplemental figure 1 and corresponding figure legend (lines 898-904)
Figure5. B) The experiment shown Gli1 mRNA levels following treatment with inhibitors of cholesterol biosynthesis, but similar findings have already been reported previously (e.g., Cooper et al, Nature Genetics 2003; Blassberg et al, Hum Mol Genet 2016), and the present results do not provide a significant conceptual advance over those earlier studies.
We thank the reviewer for this comment and for highlighting the importance of earlier studies on Hedgehog (Hh) signaling and cholesterol metabolism. While we fully agree that confirming and extending established findings has intrinsic scientific value, we respectfully disagree with the assertion that our work does not provide conceptual novelty.
The seminal work by Cooper et al. (Nature Genetics, 2003) indeed laid the foundation for linking sterol metabolism to Hedgehog signaling, and we cite it as such. However, that study was conducted in chick embryos, a model that is relatively distant from mammalian systems and human pathophysiology. Moreover, their approach relied heavily on cyclodextrin-mediated cholesterol depletion, which is non-specific and extracts multiple sterols from membranes (discussed in this article lines 512-516). In contrast, our study employs pharmacological inhibitors targeting specific enzymes in the sterol biosynthetic pathway, thereby allowing us to modulate distinct steps and intermediates in a controlled and mechanistically informative manner. We also extend these analyses to patient-derived fibroblasts and CRISPR-engineered cells, providing direct human and genetic validation of the observed effects. Importantly, we complement these cellular studies with biochemical characterization of isolated ciliary membranes from MDCK cells, enabling a direct assessment of how specific sterol alterations affect ciliary composition and Hh pathway function - an angle not addressed in prior work.
Regarding Blassberg et al. (Hum. Mol. Genet., 2016), we agree that part of our findings recapitulates their observations on SMO-related signaling defects, which we view as an important confirmation of reproducibility. However, their study primarily sought to distinguish whether Hh pathway impairment in SLOS results from 7-DHC accumulation or cholesterol depletion, concluding that cholesterol deficiency was the main cause. Our results expand on this by demonstrating that perturbations extend beyond these two sterols, and that additional intermediates in the biosynthetic pathway also impact ciliary membrane composition and signaling competence. Furthermore, our experiments using the constitutively active SmoM2 mutant show that Hh signaling defects are not restricted to SMO activation per se, revealing a broader disruption of the signaling machinery within the cilium.
Finally, neither of the above studies examined CDPX2 patient-derived cells or the consequences of EBP enzyme deficiency on Hh signaling. Our finding that this pathway is altered in this genetic context represents, to our knowledge, a novel link between CDPX2 and Hedgehog pathway dysfunction.
Taken together, our work builds upon and extends previous findings by integrating cell-type-specific, biochemical, and patient-based analyses to provide a more comprehensive and mechanistically detailed view of how sterol composition of the ciliary membrane regulates Hedgehog signaling.
In addition, the authors analyze the effect of these inhibitors on SAG stimulation, but the experiment lacks the control for Gli mRNA levels in the absence of SAG treatment. Without this control, it is impossible to know where the baseline in the experiment is and how large the effects in question really are.
Below, we provide the data expressed using the ΔΔCt method (NT + SAG normalized to NT - SAG), which more clearly illustrates the magnitude of the effect in question. As similar qPCR-based Hedgehog pathway activation assays in MEFs have been published previously (see Eguether et al., Dev. Cell 2014; Eguether et al., Mol. Biol. Cell 2018), our goal here was not to re-establish the assay itself but to highlight the comparative effects across experimental conditions. In addition, one of the datasets was obtained using a new batch of SAG, which exhibited stronger pathway activation across all conditions (visible as higher overall expression levels). To ensure valid statistical comparisons across experiments and to focus on relative rather than absolute activation, we therefore chose to present the data as fold change values, which provides a more robust and statistically consistent measure for cross-condition analysis.
J-K) The data represented in these panels for SAG treatment as fraction of Smo and its fluorescence intensity for the same sample appears to be inconsistent between the two graphs. Under SAG treatment for EBP mutants shows higher Smo fluorescence intensity while Smo positive cilia seems to be less than the wild type control cells. If the number of Smo+ cilia (quantified by eye) differs between conditions, shouldn't the quantification of Smo intensity within cilia show a similar difference?
We thank the reviewer for this careful observation. The apparent discrepancy arises because the two panels quantify different parameters. In panel (j), we counted the percentage of cilia positive for SMO (i.e., cilia in which SMO was detected above background). In contrast, panel (k) reports the fluorescence intensity of SMO, but this measurement was performed only within the SMO-positive cilia identified in panel (j). This distinction has now been explicitly clarified in the figure legend, as also suggested by Reviewer 1.
Taken together, these two analyses indicate that although fewer cilia display detectable SMO accumulation in the EBP mutant cells, the amount of SMO present within those cilia that do recruit it is comparable to wild-type levels (as reflected by the non-significant difference in fluorescence intensity). This interpretation helps explain the partial functional preservation of Hedgehog signaling in this condition and contrasts with cases such as AY9944 treatment, where both the number of SMO-positive cilia and the SMO intensity are reduced.
-
I) The rationale for using SmoM2 in the analysis of cholesterol metabolism-related diseases such as SLOS and CDPX2 is unclear. The SmoM2 variant is primarily associated with cancer rather than cholesterol biosynthesis defects and its relevance either of these disorders is not immediately apparent.
We thank the reviewer for this pertinent observation. We fully agree that SmoM2 was originally identified as an oncogenic mutation and is not directly associated with cholesterol biosynthesis disorders. However, our rationale for using this mutant was mechanistic rather than pathological. SmoM2 is a constitutively active form of SMO that triggers pathway activation independently of upstream components such as PTCH1 or ligand-mediated regulation.
By using SmoM2, we aimed to determine whether the signaling defects observed under conditions that alter sterol metabolism (e.g., treatment with AY9944 or tamoxifen) occur upstream or downstream of SMO activation. The results demonstrate that, even when SMO is constitutively active, the Hedgehog pathway remains impaired under AY9944 treatment-and to a lesser extent with tamoxifen-indicating that these sterol perturbations disrupt the pathway beyond the level of SMO activation itself. In contrast, cells treated with simvastatin maintain normal pathway responsiveness, reinforcing the specificity of this effect.
This experiment is therefore central to our study, as it reveals that sterol imbalance can hinder Hedgehog signaling even in the presence of an active SMO, providing new insight into how membrane composition influences downstream signaling competence.
Minor corrections
-
Line 385 seems to be a bit confusing which mentions cilia were treated with AY9944 - do the authors mean that cells were been treated with the drugs before isolation of cilia, or were the purified cilia actually treated with the drugs?
Thank you, this has been modified in the revised manuscript
The authors should add proper label in Figure 2 panel b for the bars representing the cilia and cell membranes.
We apologize for the oversight, the figures initially submitted with the manuscript inadvertently included some earlier versions, which explains several of the discrepancies noted by the reviewers. This issue has been corrected in the revised submission, and all figures have now been updated to reflect the finalized data.
Panels in Figure S1 should be re-arranged according to the figure legend and figure reference in line 450.
We apologize for the oversight, the figures initially submitted with the manuscript inadvertently included some earlier versions, which explains several of the discrepancies noted by the reviewers. This issue has been corrected in the revised submission, and all figures have now been updated to reflect the finalized data.
Legend for the Figure S1b should be corrected as data sets in graph represents 7 points while technical replicates in legend shows 6 experimental values.
Thank you, this has been modified in the revised manuscript
The labels for drug in Figure 3 and 5 should be corrected from tamoxifene to tamoxifen and simvastatine to simvastatin.
We apologize for the oversight, the figures initially submitted with the manuscript inadvertently included some earlier versions, which explains several of the discrepancies noted by the reviewers. This issue has been corrected in the revised submission, and all figures have now been updated to reflect the finalized data.
Reviewer #2 (Significance (Required)):
In the present study, the authors have designed a method to isolate the cilium from the MDCK cells efficiently and then utilized this procedure in conjunction with mass spectrometry to systematically analyze the sterol composition of the ciliary membrane, which they then compare to the sterol composition of the cell body. By analyzing this sterol profiling. the authors claim that the cilium has a distinct sterol composition from the cell body, including higher levels of cholesterol and desmosterol but lower levels of 8-DHC and & Lathosterol. This manuscript further demonstrates that alteration of sterol composition within cilia modulates Hedgehog signaling. These results strengthen the link between dysregulated Hedgehog signaling and defects in cholesterol biosynthesis pathways, as observed in SLOS and CDPX2.
While the ability to isolate primary cilia from cultured MDCK cells represents an important technical achievement, the central claim of the manuscript - that cilia have a different sterol composition from the cell body - is not adequately supported by the data, and more rigorous comparisons between the ciliary membrane and key organellar membranes (such as plasma membrane) are required to make this claim. Moreover, although the authors have repeatedly mention that the ciliary sterol composition is "tightly regulated" there is no evidence provided to support such claim. At best, the data suggest that the cilium and cell body may differ in sterol composition (though even that remains uncertain), but no underlying regulatory mechanisms are demonstrated. In addition, much of the 2nd half of the paper represents a rehash of experiments with sterol biosynthesis inhibitors that have already been published in the literature, making the conceptual advance modest at best. Lastly, the link between CDPX2 and defective Hedgehog signaling is tenuous.
We thank the reviewer for this detailed summary and for acknowledging the technical advance represented by our method for isolating primary cilia from MDCK cells. However, we respectfully disagree with several aspects of the reviewer's assessment of our work.
As we elaborated in our responses to earlier comments, particularly regarding Figure 5, we disagree with the characterization of part of our study as a "rehash", a somewhat derogatory word, of previously published experiments. Our approach differs from earlier studies by relying on specific pharmacological modulation of defined enzymes in the sterol biosynthesis pathway, rather than using non-specific agents such as cyclodextrins, and by linking these manipulations to direct biochemical measurements of ciliary sterol composition. This strategy allows, for the first time, a targeted and physiologically relevant examination of how specific sterol perturbations affect Hedgehog signaling.
Regarding our statement that ciliary sterol composition is "tightly regulated," we acknowledge that we have not yet explored the underlying molecular mechanisms of this regulation. Nevertheless, the experimental evidence supporting this statement lies in the variation of ciliary sterol composition across multiple treatments that strongly perturb cellular sterols. Despite broad cellular changes, the ciliary sterol profile remains very resilient for some parameters, an observation that, in our view, strongly supports the idea of a selective or regulated process maintaining ciliary sterol identity. This conclusion does not depend on comparison with other membrane compartments.
We also respectfully disagree that the observed differences between cilia and the cell body (which doesn't equal to plasma membrane) are "uncertain." The consistent enrichment in cholesterol and desmosterol, combined with the relative depletion in 8-DHC and lathosterol, were detected across independent replicates using robust lipidomic profiling and are statistically supported. These findings are, to our knowledge, the first quantitative demonstration of a sterol fingerprint specific to a mammalian cilium.
Finally, while we agree that the mechanistic link between CDPX2 and defective Hedgehog signaling warrants further exploration, the data we present, combining pharmacological inhibition (tamoxifen), CRISPR-mediated EBP knockout, and SMOM2 activation assays, all consistently indicate a functional impairment of the Hedgehog pathway under EBP deficiency. This is further reinforced by clinical reports describing Hedgehog-related phenotypes in CDPX2 patients. We therefore believe that our work provides a solid experimental and conceptual basis for connecting EBP dysfunction to Hedgehog signaling defects.
In summary, our study introduces a validated and reproducible method for mammalian cilia isolation, provides the first detailed sterol composition profile of primary cilia, and establishes a functional link between ciliary sterol imbalance and Hedgehog pathway modulation. We believe these findings represent a meaningful conceptual advance and a valuable resource for the field
Reviewer #3 (Evidence, reproducibility and clarity (Required)):
Lamaziere et al. describe an improved protocol for isolating primary cilia from MDCK cells for downstream lipidomics analysis. Using this protocol, they characterize sterol profile of MDCK cilia membrane under standard growth conditions and following pharmacological perturbations that are meant to mimic SLOS and CDPX2 disorders in humans. The authors then assess the impact of the same pharmacological manipulations on Shh pathway activity and validate their findings from these experiments using orthogonal genetic approaches. Major and minor concerns that require attention prior to publication are outlined below.
We would like to thank the reviewer for their comments
Major 1.Since the extent of contamination of the cilia preps with non-cilia membranes is unclear, and variability between replicates is not reported, it makes interpretation of changes in cilia membrane sterol composition in response to pharmacological manipulations somewhat difficult to interpret. Discussing reproducibility of cilia sterol composition between replicates (and including corresponding data) could alleviate these concerns to some extent.
We thank the reviewer for this comment. We would like to clarify that variability between replicates is indeed reported throughout the manuscript. In Figures 2 and 3, all data are presented as mean {plus minus} SEM, as indicated in the figure legends. Specifically, the data in Figure 2 are derived from six independent experiments, reflecting the central dataset used for comparative analyses, while the data in Figure 3 are based on three independent experiments.
We also note that the overall variability between replicates is low, further supporting the reproducibility of our ciliary sterol composition measurements. This consistency across independent biological replicates provides confidence that the differences observed between cilia and the cell body are robust and not due to stochastic contamination or technical variation.
2.An abundant non-ciliary membrane protein (rather than GAPDH) may be a more appropriate loading control in Fig. 1C.
This is a valuable comment and we will find a non-ciliary membrane protein to complement this experiment.
3.Fig. 2b - which bar corresponds to cells and which one to cilia? What do numbers inside bars represent? Please label accordingly.
We apologize for the oversight, the figures initially submitted with the manuscript inadvertently included some earlier versions, which explains several of the discrepancies noted by the reviewers. This issue has been corrected in the revised submission, and all figures have now been updated to reflect the finalized data.
4.Fig. 3b-d, right panels - please define what numbers inside bars represent
Thank you, this was done in the revised manuscript. The numbers are reports of absolute quantification.
5.The font in Figs 2, 3, and 4 is very small and difficult to read. Please make the font and/or panels bigger to improve readability.
We did our best to enlarge font despite space limitations, but we are willing to work with editorial staff to improve readability as suggested.
6.It would help to have a diagram of the key steps in the cholesterol synthesis pathway for reference early in the paper rather than in figure 3.
We thank the reviewer for his comment, but we don't understand why this would be helpful as we only use sterol modulators involving the pathway's enzyme in fig3. We are open to discussion with editorial staff about moving it up to fig2. If they feel this is needed
7.The authors need to discuss why/how global inhibition of enzymes (e.g. via AY9944 treatment) in a cell could cause reduction in cholesterol levels only in the cilium and not in other cell membranes (see also point 1). Yet, tamoxifen treatment lowers cholesterol across the board.
We thank the reviewer for these insightful comments. Regarding the modest overall effect of simvastatin on cholesterol levels, we would like to note that MDCK cells are an immortalized epithelial cell line with high metabolic plasticity. Such cancer-like cell types are known to exhibit enhanced de novo lipogenesis, particularly under culture conditions with ample glucose availability. This compensatory lipid biosynthesis can partially counterbalance pharmacological inhibition of the cholesterol biosynthetic pathway. Because simvastatin acts upstream in the pathway (at HMG-CoA reductase), its inhibition primarily reduces early intermediates rather than fully depleting end-product cholesterol, explaining the relatively mild changes observed in total cholesterol content. . This has been added in a new paragraph in the revised manuscript (lines 371-378).
8.Fig. 5c, g, and j - statistical analyses are missing and need to be added in support of conclusions drawn in the text of the manuscript.
Thank you, this has been done in the revised manuscript
9.The decrease in the fraction of Smo+ cilia observed in EBP KO cells is mild (panel j, no statistics), and there is possibly a clone-specific effect here as well (statistical analysis is needed to determine if EBP139 is indeed different from WT and whether EBP139 and 141 are different from each other). Similarly, Smo fluorescence intensity after SAG treatment (panel k) is the same in WT and EBP KO cells, while there is a marked difference in intraciliary Smo intensity after tamoxifen treatment. The author's conclusion "...we were able to show that results with human cells aligned with our tamoxifen experiments" (line 436) should be modified to more accurately reflect the presented data. Ditto conclusions on lines 440-442, 530-531. In fact, it is the lack of Hh phenotypes in CDPX2 patients that is consistent with the EBP KO data presented in the paper.
We thank the reviewer for this detailed comment. We have now performed the requested statistical analyses and incorporated them into the revised manuscript.
The new analyses confirm that both EBP139 and EBP141 CRISPR KO clones show a statistically significant reduction in the fraction of Smo⁺ cilia compared to WT cells. They also reveal that the two clones differ significantly from each other, consistent with the expected clonal variability inherent to independently derived CRISPR lines.
Despite this variability, several lines of evidence support our conclusion that the EBP KO phenotypes align with the effects observed after tamoxifen treatment:
1- Directionally consistent reduction in Smo⁺ cilia:
Although the magnitude of the decrease differs between clones, both clones display a significant reduction compared to WT, paralleling the reduction observed in tamoxifen-treated cells. This directional consistency is the key point for comparing pharmacological and genetic perturbations.
2-Converging evidence from SmoM2 experiments:
Tamoxifen treatment also reduces pathway output in the context of SmoM2 overexpression. This supports the interpretation that both EBP inhibition (tamoxifen) and EBP loss (CRISPR KO) impair Hedgehog signaling at the level of ciliary function, albeit more mildly than AY9944/SLOS-like perturbations.
3-Interpretation of Smo intensity (panel k):
As clarified in the revised text, the fluorescence intensities in panel K correspond only to cilia that are Smo-positive. The absence of a difference in intensity therefore does not contradict the observed reduction in the number of Smo⁺ cilia. Rather, it explains why the phenotype is milder than that observed for SLOS/AY9944: when Smo is able to enter the cilium, its enrichment level is comparable to WT.
4- Clinical relevance for CDPX2:
While Hedgehog-related phenotypes in CDPX2 patients may be milder or under-reported, several documented features, such as polydactyly (10% of cases), as well as syndactyly and clubfoot, are classically associated with ciliary/Hedgehog signaling defects. This clinical pattern is consistent with the milder yet detectable defects we observe in EBP KO cells.
Minor •Line 310: 'intraflagellar' rather than 'intraciliary' transport particle B is a more conventional term
We agree that intraflagellar is more conventional than intraciliary, but in this case, this is how the GO term is labeled in the database. In our opinion, it should stay as is.
-
Fig. 2c - typos in the color key, is grey meant to be "cells" and blue "cilia"? Individual panels are not referenced in the text
This panel has been removed thanks to comment from reviewer 1 and 3 finding it misleading.
-
Lines 357-358: "Notably, AY9944 treatment led to a greater reduction in cholesterol content as well as a greater increase in 7-DHC and 8-DHC in cilia than in the other cell membranes" - the authors need to support this statement with appropriate statistical analysis
We respectfully believe there may be a misunderstanding in the reviewer's concern. In all cases, our comparisons are made between treated vs. untreated conditions within each compartment (cell bulk vs. ciliary membrane), and the statistical significance of these differences is already reported as determined by a Mann-Whitney test. In every case, the changes observed are greater in cilia than in the cell body. The statement in the manuscript simply summarizes this quantitative observation. However, if the reviewer feels that an additional statistical test directly comparing the magnitude of the two compartment-specific changes would strengthen the claim, we are willing to include this analysis. Alternatively, if preferred, we can remove the sentence entirely, as the comparison is already clearly visible in Figure 3b.
-
Line 473 - unclear what is meant by "olfactory cilia are mainly sensory and not primary". Primary cilia are sensory.
We agree, primary cilia are sensory, but still different from cilia belonging to sensory epithelia like retina photoreceptors or olfactory cilia. Nevertheless, this statement was modified in revised manuscript
-
Line 551: 'data not shown'. Please include the data that you would like to discuss or remove discussion of these data from the manuscript.
The data is not shown because there is nothing to show, as we discussed in that sentence, use of cholesterol probe resulted in the disappearance of primary cilia altogether. We are willing to work with editorial staff to find a better way of expressing this idea.
Reviewer #3 (Significance (Required)):
Overall, the manuscript expands our knowledge of cilia membrane composition and reports an interesting link between SLOS and Shh signaling defects, which could at least in part explain SLOS patients' symptoms. The findings reported in the manuscript could be of interest to a broad audience of cell biologists and geneticists.
We would like to thank the reviewer for his recognition of the importance of this work
-
Note: This preprint has been reviewed by subject experts for Review Commons. Content has not been altered except for formatting.
Learn more at Review Commons
Referee #1
Evidence, reproducibility and clarity
Review report for 'Sterols regulate ciliary membrane dynamics and hedgehog signaling in health and disease', Lamazière et al.
In this manuscript, Lamazière et al. address an important understudied aspect of primary cilium biology, namely the sterol composition in the ciliary membrane. It is known that sterols especially play an important role in signal transduction between PTCH1 and SMO, two upstream components of the Hedgehog pathway, at the primary cilium. Moreover, several syndromes linked to cholesterol biosynthesis defects present clinical phenotypes indicative of altered Hh signal transduction. To understand the link between ciliary membrane sterol composition and Hh signal transduction in health and disease, the authors developed a method to isolate primary cilia from MDCK cells and coupled this to quantitative metabolomics. The results were validated using biophysical methods and cellular Hh signaling assays. While this is an interesting study, it is not clear from the presented data how general the findings are: can cilia be isolated from different mammalian cell types using this protocol? Is the sterol composition of MDCK cells expected to the be the same in fibroblasts or other cell types? Without this information, it is difficult to judge whether the conclusions reached in fibroblasts are indeed directly related to the sterol composition detected in MDCK cells. Below is a detailed breakdown of suggested textual changes and experimental validations to strengthen the conclusions of the manuscript.
Major comments:
- It appears that the comparison has been made between ciliary membranes and the rest of the cell's membranes, which includes many other membranes besides the plasma membrane. This significantly weakens the conclusions on the sterol content specific to the cilium, as it may in fact be highly similar to the rest of the plasma membrane. It is for example known that lathosterol is biosynthesized in the ER, and therefore the non-presence in the cilium may reflect a high abundance in the ER but not necessarily in the plasma membrane.
- While the protocol to isolate primary cilium from MDCK cells is a valuable addition to the methods available, it would be good to at least include a discussion on its general applicability. Have the authors tried to use this protocol on fibroblasts for example?
- Some of the conclusions in the introduction (lines 75-80) seem to be incorrectly phrased based on the data: in basal conditions, ciliary membranes are already enriched in cholesterol and desmosterol, and the treatment lowers this in all membranes.
- There seems to be little effect of simvastatin on overall cholesterol levels. Can the authors comment on this result? How would the membrane fluidity be altered when mimicking simvastatin-induced composition? Since the effect on Hh signaling appears to be the biggest (Figure 5B) under simvastatin treatment, it would be interesting to compare this against that found for AY9944 treatment. Also, the authors conclude that the effects of simvastatin treatment on ciliary membrane sterol composition are the mildest, however, one could argue that they are the strongest as there is a complete lack of desmosterol.
- It is not clear to me why the authors have chosen to use SAG to activate the Hh pathway, as this is a downstream mode of activation and bypasses PTCH1 (and therefore a potentially sterol-mediated interaction between the two proteins). It would be very informative to compare the effect of sterol modulation on the ability of ShhN vs SAG to activate the pathway.
- The conclusions about the effect of tamoxifen on SMO trafficking in MEFs should be validated in human patient cells before being able to conclude that there is a potential off-target effect (line 438). Also, if that is the case, the experiment of tamoxifen treatment of EBP KO cells should give an additional effect on SMO trafficking. Also, could the CDPX2 phenotypes in patients be the result of different cell types being affected than the fibroblast used in this study?
- For the experiments with the SMO-M2 mutant, it would be useful to show the extent of pathway activation by the mutant compared to SAG or ShhN treatment of non-transfected cells. Moreover, it will be necessary to exclude any direct effects of the compound treatment on the ability of this mutant to traffic to the primary cilium, which can easily be done using fluorescence microscopy as the mutant is tagged with mCherry.
Minor comments:
Line 74: 'in patients', should be rephrased to 'patient-derived cells'
Figure 2A: What do the '+/-' indicate? They seem to be erroneously placed.
Figure 2B: no label present for which bar represents cilia/other membranes
Figure 2C: this representation is slightly deceptive, since the difference between cells and cilia for lanosterol is not significantly different as shown in figure 2A.
Figure 3A: it would be useful to also show where 8-DHC is in the biosynthetic pathway.
Line 373: the title should be rephrased as it infers that DHCR7 was blocked in model membranes, which is not the case.
Lines 377-384: this paragraph seems to be a mix of methods and some explanation, but should be rephrased for clarity.
Line 403: 'which could explain the resulting defects in Hedgehog signaling': how and what defects? At this point in the study no defects in Hh signaling have been shown.
Figure 4D: 'd' is missing
Line 408: SAG treatment resulted in slightly shorter cilia: this is not the case for just SAG treated cilia, but only for the combination of SAG + AY9944. However, in that condition there appears to be a subpopulation of very short cilia, are those real?
Figure 5b: it would be good to add that all conditions contained SAG.
Figure 5D: Since it is shown in Fig 5C that there are no positive cilia -SAG, there is no point to have empty graphs in Fig 5D on the left side, nor can any statistics be done. Similarly for 5K.
Figure 5E: it is not clearly indicated what is visualized in the inserts, sometimes it's a box, sometimes a line and they seem randomly integrated into the images.
Figure 5H: is this the intensity in just SMO positive cilia? If yes, this should be indicated, and the line at '0' for WT-SAG should be removed. I am also surprised there is then ns found for WT vs SLO, since in WT there are no positive cilia, but in SLO there are a few, so it appears to be more of a black-white situation. Perhaps it would be useful to split the data from different experiments to see if it consistently the case that there is a low percentage of SMO positive cilia in SLO cells. Fig S1: panels are inverted compared to mentioning in the text.
Methods-pharmacological treatments: there appear to be large differences in concentrations chosen to treat MDCK versus MEF cells - can the authors comment on these choices and show that the enzymes are indeed inhibited at the indicated concentrations?
(optional): it would be interesting to include a gamma-tubulin staining on the cilium prep to see if there is indeed a presence of the basal body as suggested by the proteomics data.
There are many spelling mistakes and inconsistencies throughout the manuscript and its figures (mix of French and English for example) so careful proofreading would be warranted. Moreover, there are many mentionings of 'Hedgehog defects' or 'Hedgehog-linked', where in fact it is a defect in or link to the Hedgehog pathway, not the protein itself. This should be corrected.
Significance
The study of ciliary membrane composition is highly relevant to understand signal transduction in health and disease. As such, the topic of this manuscript is significant and timely. However, as indicated above, there are limitations to this study, most notably the comparison of ciliary membrane versus all cellular membranes (rather than the plasma membrane), which weakens the conclusions that can be drawn. Moreover, cell-type dependency should be more thoroughly addressed. There certainly is a methodological advance in the form of cilia isolation from MDCK cells, however, it is unclear how broadly applicable this is to other mammalian cell types.
-
-
www.reddit.com www.reddit.com
-
https://reddit.com/r/typewriters/comments/1p42tr1/typewriter_ribbon/
It's a small metal ring/hub that fits onto the ribbon spindle. You can call around to repair shops for replacements (which may be the cheapest route) https://site.xavier.edu/polt/typewriters/tw-repair.html. Ribbons Unlimited https://ribbonsunlimited.com also sells these hubs with ribbon attached, but it's more expensive to do this, but once you've got them, you can buy ribbon by itself for much cheaper in the future and just wind the new ribbon onto existing spool hubs.
Here's some useful videos which might help you out: - https://www.youtube.com/watch?v=iTFM54VKKc4 - https://www.youtube.com/watch?v=xWQTa4b7jPs (This one has some advice about using a Remington without the spools.)
-
-
teachinginhighered.com teachinginhighered.com
-
It may seem strange that I like remembering who recommended things to me, after the fact. To me, that's part of my sensemaking and ongoing relationship deepening habits.
Yes! key part of social filtering. Recommendations and source, in ones network is a key piece of metadata. I think it's not just likable but necessary.
-
-
osf.io osf.io
-
Author response:
The following is the authors’ response to the previous reviews.
Editors comments:
I would encourage you to submit a revised version that addresses the following two points:
[a] The point from Reviewer #1 about a possible major confounding factor. The following article might be germane here: Baas and Fennell, 2019: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3339568
I don’t believe that the point raised by reviewer 1 is a confounder, see my response below.
This article highlighted was in my reading list, but I did not cite it because I was confused by its methods.
The point from Reviewer #4 about the abstract. It is important that the abstract says something about how reviewers reacted to the original versions of articles in which they were cited (ie, the odds ratio = 0.84, etc result), before going on to discuss how they reacted to revised articles (ie, the odds ratio = 1.61, etc result). I would suggest doing this along the following lines - but please feel free to reword the passage "but this effect was not strong/conclusive":
When reviewers were cited in the original version of the article under review, they were less likely to approve the article compared with reviewers who were not cited, but this effect was not strong/conclusive (odds ratio = 0.84; adjusted 99.4% CI: 0.69-1.03). However, when reviewers were cited in the revised version of the article, they were more likely to approve compared with reviewers who were not cited (odds ratio = 1.61; adjusted 99.4% CI: 1.16-2.23).
I have changed the abstract to include the odds ratios for version 1 and have used the same wording as from the main text.
Reviewer #1 (Public review):
Summary:
The work used open peer reviews and followed them through a succession of reviews and author revisions. It assessed whether a reviewer had requested the author include additional citations and references to the reviewers' work. It then assessed whether the author had followed these suggestions and what the probability of acceptance was based on the authors decision. Reviewers who were cited were more likely to recommend the article for publication when compared with reviewers that were not cited. Reviewers who requested and received a citation were much likely to accept than reviewers that requested and did not receive a citation.
Strengths and weaknesses:
The work's strengths are the in-depth and thorough statistical analysis it contains and the very large dataset it uses. The methods are robust and reported in detail.
I am still concerned that there is a major confounding factor: if you ignore the reviewers requests for citations are you more likely to have ignored all their other suggestions too? This has now been mentioned briefly and slightly circuitously in the limitations section. I would still like this (I think) major limitation to be given more consideration and discussion, although I am happy that it cannot be addressed directly in the analysis.
This is likely to happen, but I do not think it’s a confounder. A confounder needs to be associated with both the outcome and the exposure of interest. If we consider forthright authors who are more likely to rebuff all suggestions, then they would receive just as many citation and self-citation requests as authors who were more compliant. The behaviour of forthright authors would likely only reduce the association seen in most authors which would be reflected in the odds ratios.
Reviewer #2 (Public review):
Summary:
This article examines reviewer coercion in the form of requesting citations to the reviewer's own work as a possible trade for acceptance and shows that, under certain conditions, this happens.
Strengths:
The methods are well done and the results support the conclusions that some reviewers "request" self-citations and may be making acceptance decisions based on whether an author fulfills that request.
Weakness:
I thank the author for addressing my comments about the original version.
Reviewer #3 (Public review):
Summary:
In this article, Barnett examines a pressing question regarding citing behavior of authors during the peer review process. In particular, the author studies the interaction between reviewers and authors, focusing on the odds of acceptance, and how this may be affected by whether or not the authors cited the reviewers' prior work, whether the reviewer requested such citations be added, and whether the authors complied/how that affected the reviewer decision-making.
Strengths:
The author uses a clever analytical design, examining four journals that use the same open peer review system, in which the identities of the authors and reviewers are both available and linkable to structured data. Categorical information about the approval is also available as structured data. This design allows a large scale investigation of this question.
Weaknesses:
My original concerns have been largely addressed. Much more detail is provided about the number of documents under consideration for each analysis, which clarifies a great deal.
Much of the observed reviewer behavior disappears or has much lower effect sizes depending on whether "Accept with Reservations" is considered an Accept or a Reject. This is acknowledged in the results text. Language has been toned down in the revised version.
The conditional analysis on the 441 reviews (lines 224-228) does support the revised interpretation as presented.
No additional concerns are noted.
Reviewer #4 (Public review):
Summary:
This work investigates whether a citation to a referee made by a paper is associated with a more positive evaluation by that referee for that paper. It provides evidence supporting this hypothesis. The work also investigates the role of self-citations by referees where the referee would ask authors to cite the referee's paper.
Strengths:
This is an important problem: referees for scientific papers must provide their impartial opinions rooted in core scientific principles. Any undue influence due to the role of citations breaks this requirement. This work studies the possible presence and extent of this.
The methods are solid and well done. The work uses a matched pair design which controls for article-level confounding and further investigates robustness to other potential confounds.
Weaknesses:
The authors have addressed most concerns in the initial review. The only remaining concern is the asymmetric reporting and highlighting of version 1 (null result) versus version 2 (rejecting null). For example the abstract says "We find that reviewers who were cited in the article under review were more likely to recommend approval, but only after the first version (odds ratio = 1.61; adjusted 99.4% CI: 1.16 to 2.23)" instead of a symmetric sentence "We find ... in version 1 and ... in version 2".
The latest version now includes the results for both versions.
Tags
Annotators
URL
-
-
www.biorxiv.org www.biorxiv.org
-
Reviewer #1 (Public review):
Summary:
This paper reports model simulations and a human behavioral experiment studying predictive learning in a multidimensional environment. The authors claim that semantic biases help people resolve ambiguity about predictive relationships due to spurious correlations.
Strengths:
(1) The general question addressed by the paper is important.
(2) The paper is clearly written.
(3) Experiments and analyses are rigorously executed.
Weaknesses:
(1) Showing that people can be misled by spurious correlations, and that they can overcome this to some extent by using semantic structure, is not especially surprising to me. Related literature already exists on illusory correlation, illusory causation, superstitious behavior, and inductive biases in causal structure learning. None of this work features in the paper, which is rather narrowly focused on a particular class of predictive representations, which, in fact, may not be particularly relevant for this experiment. I also feel that the paper is rather long and complex for what is ultimately a simple point based on a single experiment.
(2) Putting myself in the shoes of an experimental subject, I struggled to understand the nature of semantic congruency. I don't understand why the builder and terminal robots should have similar features is considered a natural semantic inductive bias. Humans build things all the time that look different from them, and we build machines that construct artifacts that look different from the machines. I think the fact that the manipulation worked attests to the ability of human subjects to pick up on patterns rather than supporting the idea that this reflects an inductive bias they brought to the experiment.
(3) As the authors note, because the experiment uses only a single transition, it's not clear that it can really test the distinctive aspects of the SR/SF framework, which come into play over longer horizons. So I'm not really sure to what extent this paper is fundamentally about SFs, as it's currently advertised.
(4) One issue with the inductive bias as defined in Equation 15 is that I don't think it will converge to the correct SR matrix. Thus, the bias is not just affecting the learning dynamics, but also the asymptotic value (if there even is one; that's not clear either). As an empirical model, this isn't necessarily wrong, but it does mess with the interpretation of the estimator. We're now talking about a different object from the SR.
(5) Some aspects of the empirical and model-based results only provide weak support for the proposed model. The following null effects don't agree with the predictions of the model:
(a) No effect of condition on reward.
(b) No effect of condition on composition spurious predictiveness.
(c) No effect of condition on the fitted bias parameter. The authors present some additional exploratory analyses that they use to support their claims, but this should be considered weaker support than the results of preregistered analyses.
(6) I appreciate that the authors were transparent about which predictions weren't confirmed. I don't think they're necessarily deal-breakers for the paper's claims. However, these caveats don't show up anywhere in the Discussion.
(7) I also worry that the study might have been underpowered to detect some of these effects. The preregistration doesn't describe any pilot data that could be used to estimate effect sizes, and it doesn't present any power analysis to support the chosen sample sizes, which I think are on the small side for this kind of study.
-
-
-
The problem is that asking the right questions requires the opposite of having zero education. You can’t just learn how to craft a prompt for an A.I. chatbot without first having the experience, exposure and, yes, education to know what the heck you are doing. The reality — and the science — is clear that learning is a messy, nonlinear human development process that resists efficiency. A.I. cannot replace it.
Why AI can't replace teachers-- "it's a human process that resists efficiency"
-
-
www.jstor.org www.jstor.orgReview1
-
"Dusty footphilosopher means the one that's poor, lives in poverty but lives in a dignified mannerand philosophizes about the universe and talks about things that well-read people talkabout, but they've never read or traveled on a plane"
the term "Dusty feet" philosophers might be an odd phrase but it's meant to represent the discovery of knowledge and insight from places that you might not expect. and in a way you can almost look at it the same way Ashanti Young explains code meshing, the "Dusty feet" philosopher is code meshing in a way, for example, African American Standard (AASE) English might look odd to someone who was taught American standard English (ASE) for years might looks at AASE and think there doing it wrong but grammatically and structurally they both follow the same rules and are both just as good, much like a "Dusty foot philosopher" who might look rough on the outside but is well educated and just as good as any other person.
-
-
drive.google.com drive.google.com
-
believe between Asian stu-dents and African Americans....We often think of this as being a Black/White issue....racism goes between every which way between all people....It’s not just one group versus another.
Although many times it can be a white-minority issue, it can also be an issue between minorities. This is why multicultural education is so important. If we are taught about different ethnic groups we’d be able to understand our differences and not let those be something that divides us but instead unites. As educators there is a responsibility to ensure that students embody a welcoming environment and contribute to a society that supports others. Multicultural education should be implemented throughout to get rid of the barriers that divide minorities and the majority from the minority.
-
-
www.biorxiv.org www.biorxiv.org
-
Note: This response was posted by the corresponding author to Review Commons. The content has not been altered except for formatting.
Learn more at Review Commons
Reply to the reviewers
We would like to thank all the reviewers for their valuable comments and criticisms. We have thoroughly revised the manuscript and the resource to address all the points raised by the reviewers. Below, we provide a point-by-point response for the sake of clarity.
Reviewer #1
__Evidence, reproducibility and clarity __
Summary: This manuscript, "MAVISp: A Modular Structure-Based Framework for Protein Variant Effects," presents a significant new resource for the scientific community, particularly in the interpretation and characterization of genomic variants. The authors have developed a comprehensive and modular computational framework that integrates various structural and biophysical analyses, alongside existing pathogenicity predictors, to provide crucial mechanistic insights into how variants affect protein structure and function. Importantly, MAVISp is open-source and designed to be extensible, facilitating reuse and adaptation by the broader community.
Major comments: - While the manuscript is formally well-structured (with clear Introduction, Results, Conclusions, and Methods sections), I found it challenging to follow in some parts. In particular, the Introduction is relatively short and lacks a deeper discussion of the state-of-the-art in protein variant effect prediction. Several methods are cited but not sufficiently described, as if prior knowledge were assumed. OPTIONAL: Extend the Introduction to better contextualize existing approaches (e.g., AlphaMissense, EVE, ESM-based predictors) and clarify what MAVISp adds compared to each.
We have expanded the introduction on the state-of-the-art of protein variant effects predictors, explaining how MAVISp departs from them.
- The workflow is summarized in Figure 1(b), which is visually informative. However, the narrative description of the pipeline is somewhat fragmented. It would be helpful to describe in more detail the available modules in MAVISp, and which of them are used in the examples provided. Since different use cases highlight different aspects of the pipeline, it would be useful to emphasize what is done step-by-step in each.
We have added a concise, narrative description of the data flow for MAVISp, as well as improved the description of modules in the main text. We will integrate the results section with a more comprehensive description of the available modules, and then clarify in the case studies which modules were applied to achieve specific results.
OPTIONAL: Consider adding a table or a supplementary figure mapping each use case to the corresponding pipeline steps and modules used.
We have added a supplementary table (Table S2) to guide the reader on the modules and workflows applied for each case study
We also added Table S1 to map the toolkit used by MAVISp to collect the data that are imported and aggregated in the webserver for further guidance.
- The text contains numerous acronyms, some of which are not defined upon first use or are only mentioned in passing. This affects readability. OPTIONAL: Define acronyms upon first appearance, and consider moving less critical technical details (e.g., database names or data formats) to the Methods or Supplementary Information. This would greatly enhance readability.
We revised the usage of acronyms following the reviewer’s directions of defying them at first appearance.
- The code and trained models are publicly available, which is excellent. The modular design and use of widely adopted frameworks (PyTorch and PyTorch Geometric) are also strong points. However, the Methods section could benefit from additional detail regarding feature extraction and preprocessing steps, especially the structural features derived from AlphaFold2 models. OPTIONAL: Include a schematic or a table summarizing all feature types, their dimensionality, and how they are computed.
We thank the reviewer for noticing and praising the availability of the tools of MAVISp. Our MAVISp framework utilizes methods and scores that incorporate machine learning features (such as EVE or RaSP), but does not employ machine learning itself. Specifically, we do not use PyTorch and do not utilize features in a machine learning sense. We do extract some information from the AlphaFold2 models that we use (such as the pLDDT score and their secondary structure content, as calculated by DSSP), and those are available in the MAVISp aggregated csv files for each protein entry and detailed in the Documentation section of the MAVISp website.
- The section on transcription factors is relatively underdeveloped compared to other use cases and lacks sufficient depth or demonstration of its practical utility. OPTIONAL: Consider either expanding this section with additional validation or removing/postponing it to a future manuscript, as it currently seems preliminary.
We have removed this section and included a mention in the conclusions as part of the future directions.
Minor comments: - Most relevant recent works are cited, including EVE, ESM-1v, and AlphaFold-based predictors. However, recent methods like AlphaMissense (Cheng et al., 2023) could be discussed more thoroughly in the comparison.
We have revised the introduction to accommodate the proper space for this comparison.
- Figures are generally clear, though some (e.g., performance barplots) are quite dense. Consider enlarging font sizes and annotating key results directly on the plots.
We have revised Figure 2 and presented only one case study to simplify its readability. We have also changed Figure 3, whereas retained the other previous figures since they seemed less problematic.
- Minor typographic errors are present. A careful proofreading is highly recommended. Below are some of the issues I identified: Page 3, line 46: "MAVISp perform" -> "MAVISp performs" Page 3, line 56: "automatically as embedded" -> "automatically embedded" Page 3, line 57: "along with to enhance" -> unclear; please revise Page 4, line 96: "web app interfaces with the database and present" -> "presents" Page 6, line 210: "to investigate wheatear" -> "whether" Page 6, lines 215-216: "We have in queue for processing with MAVISp proteins from datasets relevant to the benchmark of the PTM module." -> unclear sentence; please clarify Page 15, line 446: "Both the approaches" -> "Both approaches" Page 20, line 704: "advantage of multi-core system" -> "multi-core systems"
We have done a proofreading of the entire article, including the points above
Significance
General assessment: the strongest aspects of the study are the modularity, open-source implementation, and the integration of structural information through graph neural networks. MAVISp appears to be one of the few publicly available frameworks that can easily incorporate AlphaFold2-based features in a flexible way, lowering the barrier for developing custom predictors. Its reproducibility and transparency make it a valuable resource. However, while the technical foundation is solid and the effort substantial, the scientific narrative and presentation could be significantly improved. The manuscript is dense and hard to follow in places, with a heavy use of acronyms and insufficient explanation of key design choices. Improving the descriptive clarity, especially in the early sections, would greatly enhance the impact of this work.
Advance
to the best of my knowledge, this is one of the first modular platforms for protein variant effect prediction that integrates structural data from AlphaFold2 with bioinformatic annotations and even clinical data in an extensible fashion. While similar efforts exist (e.g., ESMfold, AlphaMissense), MAVISp distinguishes itself through openness and design for reusability. The novelty is primarily technical and practical rather than conceptual.
Audience
this study will be of strong interest to researchers in computational biology, structural bioinformatics, and genomics, particularly those developing variant effect predictors or analyzing the impact of mutations in clinical or functional genomics contexts. The audience is primarily specialized, but the open-source nature of the tool may diffuse its use among more applied or translational users, including those working in precision medicine or protein engineering.
Reviewer expertise: my expertise is in computational structural biology, molecular modeling, and (rather weak) machine learning applications in bioinformatics. I am familiar with graph-based representations of proteins, AlphaFold2, and variant effects based on Molecular Dynamics simulations. I do not have any direct expertise in clinical variant annotation pipelines.
Reviewer #2
__Evidence, reproducibility and clarity __
Summary: The authors present a pipeline and platform, MAVISp, for aggregating, displaying and analysis of variant effects with a focus on reclassification of variants of uncertain clinical significance and uncovering the molecular mechanisms underlying the mutations.
Major comments: - On testing the platform, I was unable to look-up a specific variant in ADCK1 (rs200211943, R115Q). I found that despite stating that the mapped refseq ID was NP_001136017 in the HGVSp column, it was actually mapped to the canonical UniProt sequence (Q86TW2-1). NP_001136017 actually maps to Q86TW2-3, which is missing residues 74-148 compared to the -1 isoform. The Uniprot canonical sequence has no exact RefSeq mapping, so the HGVSp column is incorrect in this instance. This mapping issue may also affect other proteins and result in incorrect HGVSp identifiers for variants.
We would like to thank the reviewer for pointing out these inconsistencies. We have revised all the entries and corrected them. If needed, the history of the cases that have been corrected can be found in the closed issues of the GitHub repository that we use for communication between biocurators and data managers (https://github.com/ELELAB/mavisp_data_collection). We have also revised the protocol we follow in this regard and the MAVISp toolkit to include better support for isoform matching in our pipelines for future entries, as well as for the revision/monitoring of existing ones, as detailed in the Method Section. In particular, we introduced a tool, uniprot2refseq, which aids the biocurator in identifying the correct match in terms of sequence length and sequence identity between RefSeq and UniProt. More details are included in the Method Section of the paper. The two relevant scripts for this step are available at: https://github.com/ELELAB/mavisp_accessory_tools/
- The paper lacks a section on how to properly interpret the results of the MAVISp platform (the case-studies are helpful, but don't lay down any global rules for interpreting the results). For example: How should a variant with conflicts between the variant impact predictors be interpreted? Are specific indicators considered more 'reliable' than others?
We have added a section in Results to clarify how to interpret results from MAVISp in the most common use cases.
- In the Methods section, GEMME is stated as being rank-normalised with 0.5 as a threshold for damaging variants. On checking the data downloaded from the site, GEMME was not rank-normalised but rather min-max normalised. Furthermore, Supplementary text S4 conflicts with the methods section over how GEMME scores are classified, S4 states that a raw-value threshold of -3 is used.
We thank the reviewer for spotting this inconsistency. This part in the main text was left over from a previous and preliminary version of the pre-print, we have revised the main text. Supplementary Text S4 includes the correct reference for the value in light of the benchmarking therewithin.
- Note. This is a major comment as one of the claims is that the associated web-tool is user-friendly. While functional, the web app is very awkward to use for analysis on any more than a few variants at once. The fixed window size of the protein table necessitates excessive scrolling to reach your protein-of-interest. This will also get worse as more proteins are added. Suggestion: add a search/filter bar. The same applies to the dataset window.
We have changed the structure of the webserver in such a way that now the whole website opens as its own separate window, instead of being confined within the size permitted by the website at DTU. This solves the fixed window size issue. Hopefully, this will improve the user experience.
We have refactored the web app by adding filtering functionality, both for the main protein table (that can now be filtered by UniProt AC, gene name or RefSeq ID) and the mutations table. Doing this required a general overhaul of the table infrastructure (we changed the underlying engine that renders the tables).
- You are unable to copy anything out of the tables.
- Hyperlinks in the tables only seem to work if you open them in a new tab or window.
The table overhauls fixed both of these issues
- All entries in the reference column point to the MAVISp preprint even when data from other sources is displayed (e.g. MAVE studies).
We clarified the meaning of the reference column in the Documentation on the MAVISp website, as we realized it had confused the reviewer. The reference column is meant to cite the papers where the computationally-generated MAVISp data are used, not external sources. Since we also have the experimental data module in the most recent release, we have also refactored the MAVISp website by adding a “Datasets and metadata” page, which details metadata for key modules. These include references to data from external sources that we include in MAVISp on a case-by-case basis (for example the results of a MAVE experiment). Additionally, we have verified that the papers using MAVISp data are updated in https://elelab.gitbook.io/mavisp/overview/publications-that-used-mavisp-data and in the csv file of the interested proteins.
Here below the current references that have been included in terms of publications using MAVISp data:
SMPD1
ASM variants in the spotlight: A structure-based atlas for unraveling pathogenic mechanisms in lysosomal acid sphingomyelinase
Biochim Biophys Acta Mol Basis Dis
38782304
https://doi.org/10.1016/j.bbadis.2024.167260
TRAP1
Point mutations of the mitochondrial chaperone TRAP1 affect its functions and pro-neoplastic activity
Cell Death & Disease
40074754
https://doi.org/10.1038/s41419-025-07467-6
BRCA2
Saturation genome editing-based clinical classification of BRCA2 variants
Nature
39779848
0.1038/s41586-024-08349-1
TP53, GRIN2A, CBFB, CALR, EGFR
TRAP1 S-nitrosylation as a model of population-shift mechanism to study the effects of nitric oxide on redox-sensitive oncoproteins
Cell Death & Disease
37085483
10.1038/s41419-023-05780-6
KIF5A, CFAP410, PILRA, CYP2R1
Computational analysis of five neurodegenerative diseases reveals shared and specific genetic loci
Computational and Structural Biotechnology Journal
38022694
https://doi.org/10.1016/j.csbj.2023.10.031
KRAS
Combining evolution and protein language models for an interpretable cancer driver mutation prediction with D2Deep
Brief Bioinform
39708841
https://doi.org/10.1093/bib/bbae664
OPTN
Decoding phospho-regulation and flanking regions in autophagy-associated short linear motifs
Communications Biology
40835742
10.1038/s42003-025-08399-9
DLG4,GRB2,SMPD1
Deciphering long-range effects of mutations: an integrated approach using elastic network models and protein structure networks
JMB
40738203
doi: 10.1016/j.jmb.2025.169359
Entering multiple mutants in the "mutations to be displayed" window is time-consuming for more than a handful of mutants. Suggestion: Add a box where multiple mutants can be pasted in at once from an external document.
During the table overhaul, we have revised the user interface to add a text box that allows free copy-pasting of mutation lists. While we understand having a single input box would have been ideal, the former selection interface (which is also still available) doesn’t allow copy-paste. This is a known limitation in Streamlit.
Minor comments
- Grammar. I appreciate that this manuscript may have been compiled by a non-native English speaker, but I would be remiss not to point out that there are numerous grammar errors throughout, usually sentence order issues or non-pluralisation. The meaning of the authors is mostly clear, but I recommend very thoroughly proof-reading the final version.
We have done proofreading on the final version of the manuscript
- There are numerous proteins that I know have high-quality MAVE datasets that are absent in the database e.g. BRCA1, HRAS and PPARG.
Yes, we are aware of this. It is far from trivial to properly import the datasets from multiplex assays. They often need to be treated on a case-by-case basis. We are in the process of carefully compiling locally all the MAVE data before releasing it within the public version of the database, so this is why they are missing. We are giving priorities to the ones that can be correlated with our predictions on changes in structural stability and then we will also cover the rest of the datasets handling them in batches. Having said this, we have checked the dataset for BRCA1, HRAS, and PPARG. We have imported the ones for PPARG and BRCA1 from ProtGym, referring to the studies published in 10.1038/ng.3700 and 10.1038/s41586-018-0461-z, respectively. Whereas for HRAS, checking in details both the available data and literature, while we did identify a suitable dataset (10.7554/eLife.27810), we struggled to understand what a sensible cut-off for discriminating between pathogenic and non-pathogenic variants would be, and so ended up not including it in the MAVISp dataset for now. We will contact the authors to clarify which thresholds to apply before importing the data.
- Checking one of the existing MAVE datasets (KRAS), I found that the variants were annotated as damaging, neutral or given a positive score (these appear to stand-in for gain-of-function variants). For better correspondence with the other columns, those with positive scores could be labelled as 'ambiguous' or 'uncertain'.
In the KRAS case study presented in MAVISP, we utilized the protein abundance dataset reported in (http://dx.doi.org/10.1038/s41586-023-06954-0) and made available in the ProteinGym repository (specifically referenced at https://github.com/OATML-Markslab/ProteinGym/blob/main/reference_files/DMS_substitutions.csv#L153). We adopted the precalculated thresholds as provided by the ProteinGym authors. In this regard, we are not really sure the reviewer is referring to this dataset or another one on KRAS.
- Numerous thresholds are defined for stabilizing / destabilizing / neutral variants in both the STABILITY and the LOCAL_INTERACTION modules. How were these thresholds determined? I note that (PMC9795540) uses a ΔΔG threshold of 1/-1 for defining stabilizing and destabilizing variants, which is relatively standard (though they also say that 2-3 would likely be better for pinpointing pathogenic variants).
We improved the description of our classification strategies for both modules in the Documentation page of our website. Also, we explained more clearly the possible sources of ‘uncertain’ annotations for the two modules in both the web app (Documentation page) and main text. Briefly, in the STABILITY module, we consider FoldX and either Rosetta or RaSP to achieve a final classification. We first classify one and the other independently, according to the following strategy:
If DDG ≥ 3, the mutation is Destabilizing If DDG ≤ −3, the mutation is Stabilizing If −2 We then compare the classifications obtained by the two methods: if they agree, then that is the final classification, if they disagree, then the final classification is Uncertain. The thresholds were selected based on a previous study, in which variants with changes in stability below 3 kcal/mol were not featuring a markedly different abundance at cellular level [10.1371/journal.pgen.1006739, 10.7554/eLife.49138]
Regarding the LOCAL_INTERACTION module, it works similarly as for the Stability module, in that Rosetta and FoldX are considered independently, and an implicit classification is performed for each, according to the rules (values in kcal/mol)
If DDG > 1, the mutation is Destabilizing. If DDG Each mutation is therefore classified for both methods. If the methods agree (i.e., if they classify the mutation in the same way), their consensus is the final classification for the mutation; if they do not agree, the final classification will be Uncertain.
If a mutation does not have an associated free energy value, the relative solvent accessible area is used to classify it: if SAS > 20%, the mutation is classified as Uncertain, otherwise it is not classified.
Thresholds here were selected according to best practices followed by the tool authors and more in general in the literature, as the reviewer also noticed.
- "Overall, with the examples in this section, we illustrate different applications of the MAVISp results, spanning from benchmarking purposes, using the experimental data to link predicted functional effects with structural mechanisms or using experimental data to validate the predictions from the MAVISp modules."
The last of these points is not an application of MAVISp, but rather a way in which external data can help validate MAVISp results. Furthermore, none of the examples given demonstrate an application in benchmarking (what is being benchmarked?).
We have revised the statements to avoid this confusion in the reader.
- Transcription factors section. This section describes an intended future expansion to MAVISp, not a current feature, and presents no results. As such, it should be moved to the conclusions/future directions section.
We have removed this section and included a mention in the conclusions as part of the future directions.
- Figures. The dot-plots generated by the web app, and in Figures 4, 5 and 6 have 2 legends. After looking at a few, it is clear that the lower legend refers to the colour of the variant on the X-axis - most likely referencing the ClinVar effect category. This is not, however, made clear either on the figures or in the app.
The reviewer’s interpretation on the second legend is correct - it does refer to the ClinVar classification. Nonetheless, we understand the positioning of the legend makes understanding what the legend refers to not obvious. We also revised the captions of the figures in the main text. On the web app, we have changed the location of the figure legend for the ClinVar effect category and added a label to make it clear what the classification refers to.
- "We identified ten variants reported in ClinVar as VUS (E102K, H86D, T29I, V91I, P2R, L44P, L44F, D56G, R11L, and E25Q, Fig.5a)" E25Q is benign in ClinVar and has had that status since first submitted.
We have corrected this in the text and the statements related to it.
Significance
Platforms that aggregate predictors of variant effect are not a new concept, for example dbNSFP is a database of SNV predictions from variant effect predictors and conservation predictors over the whole human proteome. Predictors such as CADD and PolyPhen-2 will often provide a summary of other predictions (their features) when using their platforms. MAVISp's unique angle on the problem is in the inclusion of diverse predictors from each of its different moules, giving a much wider perspective on variants and potentially allowing the user to identify the mechanistic cause of pathogenicity. The visualisation aspect of the web app is also a useful addition, although the user interface is somewhat awkward. Potentially the most valuable aspect of this study is the associated gitbook resource containing reports from biocurators for proteins that link relevant literature and analyse ClinVar variants. Unfortunately, these are only currently available for a small minority of the total proteins in the database with such reports. For improvement, I think that the paper should focus more on the precise utility of the web app / gitbook reports and how to interpret the results rather than going into detail about the underlying pipeline.
We appreciate the interest in the gitbook resource that we also see as very valuable and one of the strengths of our work. We have now implemented a new strategy based on a Python script introduced in the mavisp toolkit to generate a template Markdown file of the report that can be further customized and imported into GitBook directly (https://github.com/ELELAB/mavisp_accessory_tools/). This should allow us to streamline the production of more reports. We are currently assigning proteins in batches for reporting to biocurator through the mavisp_data_collection GitHub to expand their coverage. Also, we revised the text and added a section on the interpretation of results from MAVISp. with a focus on the utility of the web-app and reports.
In terms of audience, the fast look-up and visualisation aspects of the web-platform are likely to be of interest to clinicians in the interpretation of variants of unknown clinical significance. The ability to download the fully processed dataset on a per-protein database would be of more interest to researchers focusing on specific proteins or those taking a broader view over multiple proteins (although a facility to download the whole database would be more useful for this final group).
While our website only displays the dataset per protein, the whole dataset, including all the MAVISp entries, is available at our OSF repository (https://osf.io/ufpzm/), which is cited in the paper and linked on the MAVISp website. We have further modified the MAVISp database to add a link to the repository in the modes page, so that it is more visible.
My expertise. - I am a protein bioinformatician with a background in variant effect prediction and large-scale data analysis.
Reviewer #3 (Evidence, reproducibility and clarity (Required)):
Evidence, reproducibility and clarity:
Summary:
The authors present MAVISp, a tool for viewing protein variants heavily based on protein structure information. The authors have done a very impressive amount of curation on various protein targets, and should be commended for their efforts. The tool includes a diverse array of experimental, clinical, and computational data sources that provides value to potential users interested in a given target.
Major comments:
Unfortunately I was not able to get the website to work correctly. When selecting a protein target in simple mode, I was greeted with a completely blank page in the app window. In ensemble mode, there was no transition away from the list of targets at all. I'm using Firefox 140.0.2 (64-bit) on Ubuntu 22.04. I would like to explore the data myself and provide feedback on the user experience and utility.
We have tried reproducing the issue mentioned by the reviewer, using the exact same Ubuntu and Firefox versions, but unfortunately failed to produce it. The website worked fine for us under such an environment. The issue experienced by the reviewer may have been due to either a temporary issue with the web server or a problem with the specific browser environment they were working in, which we are unable to reproduce. It would be useful to know the date that this happened to verify if it was a downtime on the DTU IT services side that made the webserver inaccessible.
I have some serious concerns about the sustainability of the project and think that additional clarifications in the text could help. Currently is there a way to easily update a dataset to add, remove, or update a component (for example, if a new predictor is published, an error is found in a predictor dataset, or a predictor is updated)? If it requires a new round of manual curation for each protein to do this, I am worried that this will not scale and will leave the project with many out of date entries. The diversity of software tools (e.g., three different pipeline frameworks) also seems quite challenging to maintain.
We appreciate the reviewer’s concerns about long-term sustainability. It is a fair point that we consider within our steering group, who oversee and plans the activities and meet monthly. Adding entries to MAVISp is moving more and more towards automation as we grow. We aim to minimize the manual work where applicable. Still, an expert-based intervention is really needed in some of the steps, and we do not want to renounce it. We intend to keep working on MAVISp to make the process of adding and updating entries as automated as possible, and to streamline the process when manual intervention is necessary. From the point of view of the biocurators, they have three core workflows to use for the default modules, which also automatically cover the source of annotations. We are currently working to streamline the procedures behind LOCAL_INTERACTION, which is the most challenging one. On the data manager and maintainers' side, we have workflows and protocols that help us in terms of automation, quality control, etc, and we keep working to improve them. Among these, we have workflows to use for the old entries updates. As an example, the update of erroneously attributed RefSeq data (pointed out by reviewer 2) took us only one week overall (from assigning revisions and importing to the database) because we have a reduced version of Snakemake for automation that can act on only the affected modules. Also, another point is that we have streamlined the generation of the templates for the gitbook reports (see also answer to reviewer 2).
The update of old entries is planned and made regularly. We also deposit the old datasets on OSF for transparency, in case someone needs to navigate and explore the changes. We have activities planned between May and August every year to update the old entries in relation to changes of protocols in the modules, updates in the core databases that we interact with (COSMIC, Clinvar etc). In case of major changes, the activities for updates continue in the Fall. Other revisions can happen outside these time windows if an entry is needed or a specific research project and needs updates too.
Furthermore, the community of people contributing to MAVISp as biocurators or developers is growing and we have scientists contributing from other groups in relation to their research interest. We envision that for this resource to scale up, our team cannot be the only one producing data and depositing it to the database. To facilitate this we launched a pilot for a training event online (see Event page on the website) and we will repeat it once per year. We also organize regular meetings with all the active curators and developers to plan the activities in a sustainable manner and address the challenges we encounter.
As stated in the manuscript, currently with the team of people involved, automatization and resources that we have gathered around this initiative we can provide updates to the public database every third month and we have been regularly satisfied with them. Additionally, we are capable of processing from 20 to 40 proteins every month depending also on the needs of revision or expansion of analyses on existing proteins. We also depend on these data for our own research projects and we are fully committed to it.
Additionally, we are planning future activities in these directions to improve scale up and sustainability:
- Streamlining manual steps so that they are as convenient as fast as possible for our curators, e.g. by providing custom pages on the MAVISp website
- Streamline and automatize the generation of useful output, for instance the reports, by using a combination of simple automation and large language models
- Implement ways to share our software and scripts with third parties, for instance by providing ready made (or close to) containers or virtual machines
- For a future version 2 if the database grows in a direction that is not compatible with Streamlit, the web data science framework we are currently using, we will rewrite the website using a framework that would allow better flexibility and performance, for instance using Django and a proper database backend. On the same theme, according to the GitHub repository, the program relies on Python 3.9, which reaches end of life in October 2025. It has been tested against Ubuntu 18.04, which left standard support in May 2023. The authors should update the software to more modern versions of Python to promote the long-term health and maintainability of the project.
We thank the reviewer for this comment - we are aware of the upcoming EOL of Python 3.9. We tested MAVISp, both software package and web server, using Python 3.10 (which is the minimum supported version going forward) and Python 3.13 (which is the latest stable release at the time of writing) and updated the instructions in the README file on the MAVISp GitHub repository accordingly.
We plan on keeping track of Python and library versions during our testing and updating them when necessary. In the future, we also plan to deploy Continuous Integration with automated testing for our repository, making this process easier and more standardized.
I appreciate that the authors have made their code and data available. These artifacts should also be versioned and archived in a service like Zenodo, so that researchers who rely on or want to refer to specific versions can do so in their own future publications.
Since 2024, we have been reporting all previous versions of the dataset on OSF, the repository linked to the MAVISp website, at https://osf.io/ufpzm/files/osfstorage (folder: previous_releases). We prefer to keep everything under OSF, as we also use it to deposit, for example, the MD trajectory data.
Additionally, in this GitHub page that we use as a space to interact between biocurators, developers, and data managers within the MAVISp community, we also report all the changes in the NEWS space: https://github.com/ELELAB/mavisp_data_collection
Finally, the individual tools are all available in our GitHub repository, where version control is in place (see Table S1, where we now mapped all the resources used in the framework)
In the introduction of the paper, the authors conflate the clinical challenges of variant classification with evidence generation and it's quite muddled together. They should strongly consider splitting the first paragraph into two paragraphs - one about challenges in variant classification/clinical genetics/precision oncology and another about variant effect prediction and experimental methods. The authors should also note that they are many predictors other than AlphaMissense, and may want to cite the ClinGen recommendations (PMID: 36413997) in the intro instead.
We revised the introduction in light of these suggestions. We have split the paragraph as recommended and added a longer second paragraph about VEPs and using structural data in the context of VEPs. We have also added the citation that the reviewer kindly recommended.
Also in the introduction on lines 21-22 the authors assert that "a mechanistic understanding of variant effects is essential knowledge" for a variety of clinical outcomes. While this is nice, it is clearly not the case as we can classify variants according to the ACMG/AMP guidelines without any notion of specific mechanism (for example, by combining population frequency data, in silico predictor data, and functional assay data). The authors should revise the statement so that it's clear that mechanistic understanding is a worthy aspiration rather than a prerequisite.
We revised the statement in light of this comment from the reviewer
In the structural analysis section (page 5, lines 154-155 and elsewhere), the authors define cutoffs with convenient round numbers. Is there a citation for these values or were these arbitrarily chosen by the authors? I would have liked to see some justification that these assignments are reasonable. Also there seems to be an error in the text where values between -2 and -3 kcal/mol are not assigned to a bin (I assume they should also be uncertain). There are other similar seemingly-arbitrary cutoffs later in the section that should also be explained.
We have revised the text making the two intervals explicit, for better clarity.
On page 9, lines 294-298 the authors talk about using the PTEN data from ProteinGym, rather than the actual cutoffs from the paper. They get to the latter later on, but I'm not sure why this isn't first? The ProteinGym cutoffs are somewhat arbitrarily based on the median rather than expert evaluation of the dataset, and I'm not sure why it's even worth mentioning them when proper classifications are available. Regarding PTEN, it would be quite interesting to see a comparison of the VAMP-seq PTEN data and the Mighell phosphatase assay, which is cited on page 9 line 288 but is not actually a VAMP-seq dataset. I think this section could be interesting but it requires some additional attention.
We have included the data from Mighell’s phosphatase assay as provided by MAVEdb in the MAVISp database, within the experimental_data module for PTEN, and we have revised the case study, including them and explaining better the decision of supporting both the ProteinGym and MAVEdb classification in MAVISp (when available). See revised Figure3, Table 1 and corresponding text.
The authors mention "pathogenicity predictors" and otherwise use pathogenicity incorrectly throughout the manuscript. Pathogenicity is a classification for a variant after it has been curated according to a framework like the ACMG/AMP guidelines (Richards 2015 and amendments). A single tool cannot predict or assign pathogenicity - the AlphaMissense paper was wrong to use this nomenclature and these authors should not compound this mistake. These predictors should be referred to as "variant effect predictors" or similar, and they are able to produce evidence towards pathogenicity or benignity but not make pathogenicity calls themselves. For example, in Figure 4e, the terms "pathogenic" and "benign" should only be used here if these are the classifications the authors have derived from ClinVar or a similar source of clinically classified variants.
The reviewer is correct, we have revised the terminology we used in the manuscript and refers to VEPs (Variant Effect Predictors)
Minor comments:
The target selection table on the website needs some kind of text filtering option. It's very tedious to have to find a protein by scrolling through the table rather than typing in the symbol. This will only get worse as more datasets are added.
We have revised the website, adding a filtering option. In detail, we have refactored the web app by adding filtering functionality, both for the main protein table (that can now be filtered by UniProt AC, gene name, or RefSeq ID) and the mutations table. Doing this required a general overhaul of the table infrastructure (we changed the underlying engine that renders the tables).
The data sources listed on the data usage section of the website are not concordant with what is in the paper. For example, MaveDB is not listed.
We have revised and updated the data sources on the website, adding a metadata section with relevant information, including MaveDB references where applicable.
Figure 2 is somewhat confusing, as it partially interleaves results from two different proteins. This would be nicer as two separate figures, one on each protein, or just of a single protein.
As suggested by the reviewer, we have now revised the figure and corresponding legends and text, focusing only on one of the two proteins.
Figure 3 panel b is distractingly large and I wonder if the authors could do a little bit more with this visualization.
We have revised Figure 3 to solve these issues and integrating new data from the comparison with the phosphatase assay
Capitalization is inconsistent throughout the manuscript. For example, page 9 line 288 refers to VampSEQ instead of VAMP-seq (although this is correct elsewhere). MaveDB is referred to as MAVEdb or MAVEDB in various places. AlphaMissense is referred to as Alphamissense in the Figure 5 legend. The authors should make a careful pass through the manuscript to address this kind of issues.
We have carefully proofread the paper for these inconsistencies
MaveDB has a more recent paper (PMID: 39838450) that should be cited instead of/in addition to Esposito et al.
We have added the reference that the reviewer recommended
On page 11, lines 338-339 the authors mention some interesting proteins including BLC2, which has base editor data available (PMID: 35288574). Are there plans to incorporate this type of functional assay data into MAVISp?
The assay mentioned in the paper refers to an experimental setup designed to investigate mutations that may confer resistance to the drug venetoclax. We started the first steps to implement a MAVISp module aimed at evaluating the impact of mutations on drug binding using alchemical free energy perturbations (ensemble mode) but we are far from having it complete. We expect to import these data when the module will be finalized since they can be used to benchmark it and BCL2 is one of the proteins that we are using to develop and test the new module.
Reviewer #3 (Significance (Required)):
Significance:
General assessment:
This is a nice resource and the authors have clearly put a lot of effort in. They should be celebrated for their achievments in curating the diverse datasets, and the GitBooks are a nice approach. However, I wasn't able to get the website to work and I have raised several issues with the paper itself that I think should be addressed.
Advance:
New ways to explore and integrate complex data like protein structures and variant effects are always interesting and welcome. I appreciate the effort towards manual curation of datasets. This work is very similar in theme to existing tools like Genomics 2 Proteins portal (PMID: 38260256) and ProtVar (PMID: 38769064). Unfortunately as I wasn't able to use the site I can't comment further on MAVISp's position in the landscape.
We have expanded the conclusions section to add a comparison and cite previously published work, and linked to a review we published last year that frames MAVISp in the context of computational frameworks for the prediction of variant effects. In brief, the Genomics 2 Proteins portal (G2P) includes data from several sources, including some overlapping with MAVISp such as Phosphosite or MAVEdb, as well as features calculated on the protein structure. ProtVar also aggregates mutations from different sources and includes both variant effect predictors and predictions of changes in stability upon mutation, as well as predictions of complex structures. These approaches are only partially overlapping with MAVISp. G2P is primarily focused on structural and other annotations of the effect of a mutation; it doesn’t include features about changes of stability, binding, or long-range effects, and doesn’t attempt to classify the impact of a mutation according to its measurements. It also doesn’t include information on protein dynamics. Similarly, ProtVar does include information on binding free energies, long effects, or dynamical information.
Audience:
MAVISp could appeal to a diverse group of researchers who are interested in the biology or biochemistry of proteins that are included, or are interested in protein variants in general either from a computational/machine learning perspective or from a genetics/genomics perspective.
My expertise:
I am an expert in high-throughput functional genomics experiments and am an experienced computational biologist with software engineering experience.
-
Note: This preprint has been reviewed by subject experts for Review Commons. Content has not been altered except for formatting.
Learn more at Review Commons
Referee #3
Evidence, reproducibility and clarity
Summary:
The authors present MAVISp, a tool for viewing protein variants heavily based on protein structure information. The authors have done a very impressive amount of curation on various protein targets, and should be commended for their efforts. The tool includes a diverse array of experimental, clinical, and computational data sources that provides value to potential users interested in a given target.
Major comments:
Unfortunately I was not able to get the website to work properly. When selecting a protein target in simple mode, I was greeted with a completely blank page in the app window, and in ensemble mode, there was no transition away from the list of targets at all. I'm using Firefox 140.0.2 (64-bit) on Ubuntu 22.04. I would have liked to be able to explore the data myself and provide feedback on the user experience and utility.
I have some serious concerns about the sustainability of the project and think that additional clarifications in the text could help. Currently is there a way to easily update a dataset to add, remove, or update a component (for example, if a new predictor is published, an error is found in a predictor dataset, or a predictor is updated)? If it requires a new round of manual curation for each protein to do this, I am worried that this will not scale and will leave the project with many out of date entries. The diversity of software tools (e.g., three different pipeline frameworks) also seems quite challenging to maintain.
On the same theme, according to the GitHub repository, the program relies on Python 3.9, which reaches end of life in October 2025. It has been tested against Ubuntu 18.04, which left standard support in May 2023. The authors should update the software to more modern versions of Python to promote the long-term health and maintainability of the project.
I appreciate that the authors have made their code and data available. These artifacts should also be versioned and archived in a service like Zenodo, so that researchers who rely on or want to refer to specific versions can do so in their own future publications.
In the introduction of the paper, the authors conflate the clinical challenges of variant classification with evidence generation and it's quite muddled together. The y should strongly consider splitting the first paragraph into two paragraphs - one about challenges in variant classification/clinical genetics/precision oncology and another about variant effect prediction and experimental methods. The authors should also note that they are many predictors other than AlphaMissense, and may want to cite the ClinGen recommendations (PMID: 36413997) in the intro instead.
Also in the introduction on lines 21-22 the authors assert that "a mechanistic understanding of variant effects is essential knowledge" for a variety of clinical outcomes. While this is nice, it is clearly not the case as we are able to classify variants according to the ACMG/AMP guidelines without any notion of specific mechanism (for example, by combining population frequency data, in silico predictor data, and functional assay data). The authors should revise the statement so that it's clear that mechanistic understanding is a worthy aspiration rather than a prerequisite.
In the structural analysis section (page 5, lines 154-155 and elsewhere), the authors define cutoffs with convenient round numbers. Is there a citation for these values or were these arbitrarily chosen by the authors? I would have liked to see some justification that these assignments are reasonable. Also there seems to be an error in the text where values between -2 and -3 kcal/mol are not assigned to a bin (I assume they should also be uncertain). There are other similar seemingly-arbitrary cutoffs later in the section that should also be explained.
On page 9, lines 294-298 the authors talk about using the PTEN data from ProteinGym, rather than the actual cutoffs from the paper. They get to the latter later on, but I'm not sure why this isn't first? The ProteinGym cutoffs are somewhat arbitrarily based on the median rather than expert evaluation of the dataset and I'm not sure why it's even worth mentioning them when proper classifications are available. Regarding PTEN, it would be quite interesting to see a comparison of the VAMP-seq PTEN data and the Mighell phosphatase assay, which is cited on page 9 line 288 but is not actually a VAMP-seq dataset. I think this section could be interesting but it requires some additional attention.
The authors mention "pathogenicity predictors" and otherwise use pathogenicity incorrectly throughout the manuscript. Pathogenicity is a classification for a variant after it has been curated according to a framework like the ACMG/AMP guidelines (Richards 2015 and amendments). A single tool cannot predict or assign pathogenicity - the AlphaMissense paper was wrong to use this nomenclature and these authors should not compound this mistake. These predictors should be referred to as "variant effect predictors" or similar, and they are able to produce evidence towards pathogenicity or benignity but not make pathogenicity calls themselves. For example, in Figure 4e, the terms "pathogenic" and "benign" should only be used here if these are the classifications the authors have derived from ClinVar or a similar source of clinically classified variants.
Minor comments:
The target selection table on the website needs some kind of text filtering option. It's very tedious to have to find a protein by scrolling through the table rather than typing in the symbol. This will only get worse as more datasets are added.
The data sources listed on the data usage section of the website are not concordant with what is in the paper. For example, MaveDB is not listed.
I found Figure 2 to be a bit confusing in that it partially interleaves results from two different proteins. I think this would be nicer as two separate figures, one on each protein, or just of a single protein.
Figure 3 panel b is distractingly large and I wonder if the authors could do a little bit more with this visualization.
Capitalization is inconsistent throughout the manuscript. For example, page 9 line 288 refers to VampSEQ instead of VAMP-seq (although this is correct elsewhere). MaveDB is referred to as MAVEdb or MAVEDB in various places. AlphaMissense is referred to as Alphamissense in the Figure 5 legend. The authors should make a careful pass through the manuscript to address this kind of issues.
MaveDB has a more recent paper (PMID: 39838450) that should be cited instead of/in addition to Esposito et al.
On page 11, lines 338-339 the authors mention some interesting proteins including BLC2, which has base editor data available (PMID: 35288574). Are there plans to incorporate this type of functional assay data into MAVISp?
Significance
General assessment:
This is a nice resource and the authors have clearly put a lot of effort in. They should be celebrated for their achievments in curating the diverse datasets, and the GitBooks are a nice approach. However, I wasn't able to get the website to work and I have raised several issues with the paper itself that I think should be addressed.
Advance:
New ways to explore and integrate complex data like protein structures and variant effects are always interesting and welcome. I appreciate the effort towards manual curation of datasets. This work is very similar in theme to existing tools like Genomics 2 Proteins portal (PMID: 38260256) and ProtVar (PMID: 38769064). Unfortunately as I wasn't able to use the site I can't comment further on MAVISp's position in the landscape.
Audience:
MAVISp could appeal to a diverse group of researchers who are interested in the biology or biochemistry of proteins that are included, or are interested in protein variants in general either from a computational/machine learning perspective or from a genetics/genomics perspective.
My expertise:
I am an expert in high-throughput functional genomics experiments and am an experienced computational biologist with software engineering experience.
-
-
inst-fs-iad-prod.inscloudgate.net inst-fs-iad-prod.inscloudgate.netview1
-
We define multicultural education in a sociopolitical context as follows: Multi-cultural education is a process of comprehensive school reform and basic education for all students. It challenges and rejects racism and other forms of discrimination in schools and society and accepts and affirms the pluralism (ethnic, racial, linguistic, religious, economic, and gender, among others) that stu-dents, their communities, and teachers reflect. Multicultural education permeates schools' curriculum and instructional strategies as well as the interactions among teachers, students, and families and the very way that schools conceptualize the nature of teaching and learning. Because it uses critical pedagogy as its under-lying philosophy and focuses on knowledge, reflection, and action (praxis) as the basis for social change, multicultural education promotes democratic principles of social justice.
This definition is powerful because it frames multicultural education as active anti-racism, not just passive celebration. Calling it a "process" and "critical pedagogy" means the goal isn't just knowledge, but action for social justice. This is a radical idea—it's not about adding a unit, but about fundamentally questioning how schools operate and whose knowledge is valued. The real challenge is moving from this ideal to practice in a system that often prefers the safer, superficial versions. BUT this often leads to the neglect of the aspects that we actually need to pay attention to.
-
-
www.biorxiv.org www.biorxiv.org
-
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public Review):
Summary:
In this study, the authors trained a variational autoencoder (VAE) to create a high-dimensional "voice latent space" (VLS) using extensive voice samples, and analyzed how this space corresponds to brain activity through fMRI studies focusing on the temporal voice areas (TVAs). Their analyses included encoding and decoding techniques, as well as representational similarity analysis (RSA), which showed that the VLS could effectively map onto and predict brain activity patterns, allowing for the reconstruction of voice stimuli that preserve key aspects of speaker identity.
Strengths:
This paper is well-written and easy to follow. Most of the methods and results were clearly described. The authors combined a variety of analytical methods in neuroimaging studies, including encoding, decoding, and RSA. In addition to commonly used DNN encoding analysis, the authors performed DNN decoding and resynthesized the stimuli using VAE decoders. Furthermore, in addition to machine learning classifiers, the authors also included human behavioral tests to evaluate the reconstruction performance.
Weaknesses:
This manuscript presents a variational autoencoder (VAE) to evaluate voice identity representations from brain recordings. However, the study's scope is limited by testing only one model, leaving unclear how generalizable or impactful the findings are. The preservation of identity-related information in the voice latent space (VLS) is expected, given the VAE model's design to reconstruct original vocal stimuli. Nonetheless, the study lacks a deeper investigation into what specific aspects of auditory coding these latent dimensions represent. The results in Figure 1c-e merely tested a very limited set of speech features. Moreover, there is no analysis of how these features and the whole VAE model perform in standard speech tasks like speech recognition or phoneme recognition. It is not clear what kind of computations the VAE model presented in this work is capable of. Inclusion of comparisons with state-of-the-art unsupervised or self-supervised speech models known for their alignment with auditory cortical responses, such as Wav2Vec2, HuBERT, and Whisper, would strengthen the validation of the VAE model and provide insights into its relative capabilities and limitations.
The claim that the VLS outperforms a linear model (LIN) in decoding tasks does not significantly advance our understanding of the underlying brain representations. Given the complexity of auditory processing, it is unsurprising that a nonlinear model would outperform a simpler linear counterpart. The study could be improved by incorporating a comparative analysis with alternative models that differ in architecture, computational strategies, or training methods. Such comparisons could elucidate specific features or capabilities of the VLS, offering a more nuanced understanding of its effectiveness and the computational principles it embodies. This approach would allow the authors to test specific hypotheses about how different aspects of the model contribute to its performance, providing a clearer picture of the shared coding in VLS and the brain.
The manuscript overlooks some crucial alternative explanations for the discriminant representation of vocal identity. For instance, the discriminant representation of vocal identity can be either a higher-level abstract representation or a lower-level coding of pitch height. Prior studies using fMRI and ECoG have identified both types of representation within the superior temporal gyrus (STG) (e.g., Tang et al., Science 2017; Feng et al., NeuroImage 2021). Additionally, the methodology does not clarify whether the stimuli from different speakers contained identical speech content. If the speech content varied across speakers, the approach of averaging trials to obtain a mean vector for each speaker-the "identity-based analysis"-may not adequately control for confounding acoustic-phonetic features. Notably, the principal component 2 (PC2) in Figure 1b appears to correlate with absolute pitch height, suggesting that some aspects of the model's effectiveness might be attributed to simpler acoustic properties rather than complex identity-specific information.
Methodologically, there are issues that warrant attention. In characterizing the autoencoder latent space, the authors initialized logistic regression classifiers 100 times and calculated the tstatistics using degrees of freedom (df) of 99. Given that logistic regression is a convex optimization problem typically converging to a global optimum, these multiple initializations of the classifier were likely not entirely independent. Consequently, the reported degrees of freedom and the effect size estimates might not accurately reflect the true variability and independence of the classifier outcomes. A more careful evaluation of these aspects is necessary to ensure the statistical robustness of the results.
We thank Reviewer #1 for their thoughtful and constructive comments. Below, we address the key points raised:
New comparitive models. We agree there are still many open questions on the structure of the VLS and the specific aspects of auditory coding that its latent dimensions represent. The features tested in Figure 1c-e are not speech features, but aspects related to speaker identity: age, gender and unique identity. Nevertheless we agree the VLS could be compared to recent speech models (not available when we started this project): we have now included comparisons with Wav2Vec and HuBERT in the encoding section (new Figure 2-S3). The comparison of encoding results based on LIN, the VLS, Wav2Vec and HuBERT (new Fig2S3) indicates no clear superiority of one model over the others; rather, different sets of voxels are better explained by the different models. Interestingly all four models yielded best encoding results for the m and a TVA, indicating some consistency across models.
On decoding directly from spectrograms. We have now added decoding results obtained directly from spectrograms, as requested in the private review. These are presented in the revised Figure 4, and allow for comparison with the LIN- and VLS-based reconstructions. As noted, spectrogram-based reconstructions sounded less vocal-like and faithful to the original, confirming that the latent spaces capture more abstract and cerebral-like voice representations.
On the number and length of stimuli. The rationale for using a large number of brief, randomly spliced speech excerpts from different languages was to extract identity features independent of specific linguistic cues. Indeed, the PC2 could very well correlate with pitch; we were not able to extract reliable f0 information from the thousands of brief stimuli, many of which are largely inharmonic (e.g., fricatives), such that this assumption could not be tested empirically. But it would be relevant that the weight of PC2 correlates with pitch: although the average fundamental frequency of phonation is not a linguistic cue, it is a major acoustical feature differentiating speaker identities.
Statistics correction. To address the issue of potential dependence between multiple runs of logistic regression, we replaced our previous analysis with a Wilcoxon signedrank test comparing decoding accuracies to chance. The results remain significant across classifications, and the revised figure and text reflect this change.
Reviewer #2 (Public Review):
Summary:
Lamothe et al. collected fMRI responses to many voice stimuli in 3 subjects. The authors trained two different autoencoders on voice audio samples and predicted latent space embeddings from the fMRI responses, allowing the voice spectrograms to be reconstructed. The degree to which reconstructions from different auditory ROIs correctly represented speaker identity, gender, or age was assessed by machine classification and human listener evaluations. Complementing this, the representational content was also assessed using representational similarity analysis. The results broadly concur with the notion that temporal voice areas are sensitive to different types of categorical voice information.
Strengths:
The single-subject approach that allows thousands of responses to unique stimuli to be recorded and analyzed is powerful. The idea of using this approach to probe cortical voice representations is strong and the experiment is technically solid.
Weaknesses:
The paper could benefit from more discussion of the assumptions behind the reconstruction analyses and the conclusions it allows. The authors write that reconstruction of a stimulus from brain responses represents 'a robust test of the adequacy of models of brain activity' (L138). I concur that stimulus reconstruction is useful for evaluating the nature of representations, but the notion that they can test the adequacy of the specific autoencoder presented here as a model of brain activity should be discussed at more length. Natural sounds are correlated in many feature dimensions and can therefore be summarized in several ways, and similar information can be read out from different model representations. Models trained to reconstruct natural stimuli can exploit many correlated features and it is quite possible that very different models based on different features can be used for similar reconstructions. Reconstructability does not by itself imply that the model is an accurate brain model. Non-linear networks trained on natural stimuli are arguably not tested in the same rigorous manner as models built to explicitly account for computations (they can generate predictions and experiments can be designed to test those predictions). While it is true that there is increasing evidence that neural network embeddings can predict brain data well, it is still a matter of debate whether good predictability by itself qualifies DNNs as 'plausible computational models for investigating brain processes' (L72). This concern is amplified in the context of decoding and naturalistic stimuli where many correlated features can be represented in many ways. It is unclear how much the results hinge on the specificities of the specific autoencoder architectures used. For instance, it would be useful to know the motivations for why the specific VAE used here should constitute a good model for probing neural voice representations.
Relatedly, it is not clear how VAEs as generative models are motivated as computational models of voice representations in the brain. The task of voice areas in the brain is not to generate voice stimuli but to discriminate and extract information. The task of reconstructing an input spectrogram is perhaps useful for probing information content, but discriminative models, e.g., trained on the task of discriminating voices, would seem more obvious candidates. Why not include discriminatively trained models for comparison?
The autoencoder learns a mapping from latent space to well-formed voice spectrograms. Regularized regression then learns a mapping between this latent space and activity space. All reconstructions might sound 'natural', which simply means that the autoencoder works. It would be good to have a stronger test of how close the reconstructions are to the original stimulus. For instance, is the reconstruction the closest stimulus to the original in latent space coordinates out of using the experimental stimuli, or where does it rank? How do small changes in beta amplitudes impact the reconstruction? The effective dimensionality of the activity space could be estimated, e.g. by PCA of the voice samples' contrast maps, and it could then be estimated how the main directions in the activity space map to differences in latent space. It would be good to get a better grasp of the granularity of information that can be decoded/ reconstructed.
What can we make of the apparent trend that LIN is higher than VLS for identity classification (at least VLS does not outperform LIN)? A general argument of the paper seems to be that VLS is a better model of voice representations compared to LIN as a 'control' model. Then we would expect VLS to perform better on identity classification. The age and gender of a voice can likely be classified from many acoustic features that may not require dedicated voice processing.
The RDM results reported are significant only for some subjects and in some ROIs. This presumably means that results are not significant in the other subjects. Yet, the authors assert general conclusions (e.g. the VLS better explains RDM in TVA than LIN). An assumption typically made in single-subject studies (with large amounts of data in individual subjects) is that the effects observed and reported in papers are robust in individual subjects. More than one subject is usually included to hint that this is the case. This is an intriguing approach. However, reports of effects that are statistically significant in some subjects and some ROIs are difficult to interpret. This, in my view, runs contrary to the logic and leverage of the single-subject approach. Reporting results that are only significant in 1 out of 3 subjects and inferring general conclusions from this seems less convincing.
The first main finding is stated as being that '128 dimensions are sufficient to explain a sizeable portion of the brain activity' (L379). What qualifies this? From my understanding, only models of that dimensionality were tested. They explain a sizeable portion of brain activity, but it is difficult to follow what 'sizable' is without baseline models that estimate a prediction floor and ceiling. For instance, would autoencoders that reconstruct any spectrogram (not just voice) also predict a sizable portion of the measured activity? What happens to reconstruction results as the dimensionality is varied?
A second main finding is stated as being that the 'VLS outperforms the LIN space' (L381). It seems correct that the VAE yields more natural-sounding reconstructions, but this is a technical feature of the chosen autoencoding approach. That the VLS yields a 'more brain-like representational space' I assume refers to the RDM results where the RDM correlations were mainly significant in one subject. For classification, the performance of features from the reconstructions (age/ gender/ identity) gives results that seem more mixed, and it seems difficult to draw a general conclusion about the VLS being better. It is not clear that this general claim is well supported.
It is not clear why the RDM was not formed based on the 'stimulus GLM' betas. The 'identity GLM' is already biased towards identity and it would be stronger to show associations at the stimulus level.
Multiple comparisons were performed across ROIs, models, subjects, and features in the classification analyses, but it is not clear how correction for these multiple comparisons was implemented in the statistical tests on classification accuracies.
Risks of overfitting and bias are a recurrent challenge in stimulus reconstruction with fMRI. It would be good with more control analyses to ensure that this was not the case. For instance, how were the repeated test stimuli presented? Were they intermingled with the other stimuli used for training or presented in separate runs? If intermingled, then the training and test data would have been preprocessed together, which could compromise the test set. The reconstructions could be performed on responses from independent runs, preprocessed separately, as a control. This should include all preprocessing, for instance, estimating stimulus/identity GLMs on separately processed run pairs rather than across all runs. Also, it would be good to avoid detrending before GLM denoising (or at least testing its effects) as these can interact.
We appreciate Reviewer #2’s careful reading and numerous suggestions for improving clarity and presentation. We have implemented the suggested text edits, corrected ambiguities, and clarified methodological details throughout the manuscript. In particular, we have toned down several sentences that we agree were making strong claims (L72, L118, L378, L380-381).
Clarifications, corrections and additional information:
We streamlined the introduction by reducing overly specific details and better framing the VLS concept before presenting specifics.
Clarified the motivation for the age classification split and corrected several inaccuracies and ambiguities in the methods, including the hearing thresholds, balancing of category levels, and stimulus energy selection procedure.
Provided additional information on the temporal structure of runs and experimental stimuli selection.
Corrected the description of technical issues affecting one participant and ensured all acronyms are properly defined in the text and figure legends.
Confirmed that audiograms were performed repeatedly to monitor hearing thresholds and clarified our use of robust scaling and normalization procedures.
Regarding the test of RDM correlations, we clarified in the text that multiple comparisons were corrected using a permutation-based framework.
Reviewer #3 (Public Review):
Summary:
In this manuscript, Lamothe et al. sought to identify the neural substrates of voice identity in the human brain by correlating fMRI recordings with the latent space of a variational autoencoder (VAE) trained on voice spectrograms. They used encoding and decoding models, and showed that the "voice" latent space (VLS) of the VAE performs, in general, (slightly) better than a linear autoencoder's latent space. Additionally, they showed dissociations in the encoding of voice identity across the temporal voice areas.
Strengths:
The geometry of the neural representations of voice identity has not been studied so far. Previous studies on the content of speech and faces in vision suggest that such geometry could exist. This study demonstrates this point systematically, leveraging a specifically trained variational autoencoder.
The size of the voice dataset and the length of the fMRI recordings ensure that the findings are robust.
Weaknesses:
Overall, the VLS is often only marginally better than the linear model across analysis, raising the question of whether the observed performance improvements are due to the higher number of parameters trained in the VAE, rather than the non-linearity itself. A fair comparison would necessitate that the number of parameters be maintained consistently across both models, at least as an additional verification step.
The encoding and RSM results are quite different. This is unexpected, as similar embedding geometries between the VLS and the brain activations should be reflected by higher correlation values of the encoding model.
The consistency across participants is not particularly high, for instance, S1 seemed to have demonstrated excellent performances, while S2 showed poor performance.
An important control analysis would be to compare the decoding results with those obtained by a decoder operating directly on the latent spaces, in order to further highlight the interest of the non-linear transformations of the decoder model. Currently, it is unclear whether the non-linearity of the decoder improves the decoding performance, considering the poor resemblance between the VLS and brain-reconstructed spectrograms.
We thank Reviewer #3 for their comments. In response:
Code and preprocessed data are now available as indicated in the revised manuscript.
While we appreciate the suggestion to display supplementary analyses as boxplots split by hemisphere, we opted to retain the current format as we do not have hypotheses regarding hemispheric lateralization, and the small sample size per hemisphere would preclude robust conclusions.
Confirmed that the identities in Figure 3a are indeed ordered by age and have clarified this in the legend.
The higher variance observed in correlations for the aTVA in Figure 3b reflects the small number of data points (3 participants × 2 hemispheres), and this is now explained.
Regarding the cerebral encoding of gender and age, we acknowledge this interesting pattern. Prior work (e.g., Charest et al., 2013) found overlapping processing regions for voice gender without clear subregional differences in the TVAs. Evidence on voice age encoding remains sparse, and we highlight this novel finding in our discussion.
We again thank the reviewers for their insightful comments, which have greatly improved the quality and clarity of our work.
Reviewer #1 (Recommendations For The Authors):
(1) A set of recent advances have shown that embeddings of unsupervised/self-supervised speech models aligned to auditory responses to speech in the temporal cortex (e.g. Wav2Vec2: Millet et al NeurIPS 2022; HuBERT: Li et al. Nat Neurosci 2023; Whisper: Goldstein et al.bioRxiv 2023). These models are known to preserve a variety of speech information (phonetics, linguistic information, emotions, speaker identity, etc) and perform well in a variety of downstream tasks. These other models should be evaluated or at least discussed in the study.
We fully agree - the pace of progress in this area of voice technology has been incredible. Many of these models were not yet available at the time this work started so we could not use them in our comparison with cerebral representations.
We have now implemented Reviewer #1’s suggestion and evaluated Wav2Vec and HuBERT. The results are presented in supplementary Figure 2-S3. Correlations between activity predicted by the model and the real activity were globally comparable with those obtained with the LIN and VLS models. Interestingly both HuBERT and Wav2Vec yielded highest correlations in the mTVA, and to a lesser extent, the aTVA, as the LIN and VLS models.
(2) The test statistics of the results in Fig 1c-e need to be revised. Given that logistic regression is a convex optimization problem typically converging to a global optimum, these multiple initializations of the classifier were likely not entirely independent. Consequently, the reported degrees of freedom and the effect size estimates might not accurately reflect the true variability and independence of the classifier outcomes. A more careful evaluation of these aspects is necessary to ensure the statistical robustness of the results.
We thank Reviewer #1 for pointing out this important issue regarding the potential dependence between multiple runs of the logistic regression model. To address this concern, we have revised our analyses and used a Wilcoxon signed-rank test to compare the decoding accuracy to chance level. The results showed that the accuracy was significantly above chance for all classifications (Wilcoxon signed-rank test, all W=15, p=0.03125). We updated Figure 1c-e and the corresponding text (L154-L155) to reflect the revised analysis. Because the focus of this section is to probe the informational content of the autoencoder’s latent spaces, and since there are only 5 decoding accuracy values per model, we dropped the inter-model statistical test.
(3) In Line 198, the authors discuss the number of dimensions used in their models. To provide a comprehensive comparison, it would be informative to include direct decoding results from the original spectrograms alongside those from the VLS and LIN models. Given the vast diversity in vocal speech characteristics, it is plausible that the speaker identities might correlate with specific speech-related features also represented in both the auditory cortex and the VLS. Therefore, a clearer understanding of the original distribution of voice identities in the untransformed auditory space would be beneficial. This addition would help ascertain the extent to which transformations applied by the VLS or LIN models might be capturing or obscuring relevant auditory information.
We have now implemented Reviewer #1’s suggestion. The graphs on the right panel b of revised Figure 4 now show decoding results obtained from the regression performed directly on the spectrograms, rather than on representations of them, for our two example test stimuli. They can be listened to and compared to the LIN- and VLS-based reconstructions in Supplementary Audio 2. Compared to the LIN and VLS, the SPEC-based reconstructions sounded much less vocal or similar to the original, indicating that the latent spaces indeed capture more abstract voice representations, more similar to cerebral ones.
Reviewer #2 (Recommendations For The Authors):
L31: 'in voice' > consider rewording (from a voice?).
L33: consider splitting sentence (after interactions).
L39: 'brain' after parentheses.
L45-: certainly DNNs 'as a powerful tool' extend to audio (not just image and video) beyond their use in brain models.
L52: listened to / heard.
L63: use second/s consistently.
L64: the reference to Figure 5D is maybe a bit confusing here in the introduction.
We thank Reviewer #2 for these recommendations, which we have implemented.
L79-88: this section is formulated in a way that is too detailed for the introduction text (confusing to read). Consider a more general introduction to the VLS concept here and the details of this study later.
L99-: again, I think the experimental details are best saved for later. It's good to provide a feel for the analysis pipeline here, but some of the details provided (number of averages, denoising, preprocessing), are anyway too unspecific to allow the reader to fully follow the analysis.
Again, thank you for these suggestions for improving readability: we have modified the text accordingly.
L159: what was the motivation for classifying age as a 2-class classification problem? Rather than more classes or continuous prediction? How did you choose the age split?
The motivation for the 2 age classes was to align on the gender classification task for better comparison. The cutoff (30 years) was not driven by any scientific consideration, but by practical ones, based on the median age in our stimulus set. This is now clarified in the manuscript (L149).
L263: Is the test of RDM correlation>0 corrected for multiple comparisons across ROIs, subjects, and models?
The test of RDM correlation>0 was indeed corrected for multiple comparisons for models using the permutation-based ‘maximum statistics’ framework for multiple comparison correction (described in Giordano et al., 2023 and Maris & Oostenveld, 2007). This framework was applied for each ROI and subject. It was described in the Methods (L745) but not clearly enough in the text—we thank Reviewer #2 and clarified it in the text (L246, L260-L261).
L379: 'these stimuli' - weren't the experimental stimuli different from those used to train the V/AE?
We thank Reviewer #2 for spotting this issue. Indeed, the experimental stimuli are different from those used to train the models. We corrected the text to reflect this distinction (L84-L85).
L443: what are 'technical issues' that prevented subject 3 from participating in 48 runs??
We thank Reviewer #2 for pointing out the ambiguity in our previous statement. Participant 3 actually experienced personal health concerns that prevented them from completing the whole number of runs. We corrected this to provide a more accurate description (L442-L443).
L444: participants were instructed to 'stay in the scanner'!? Do you mean 'stay still', or something?
We thank the Reviewer for spotting this forgotten word. We have corrected the passage (L444).
L463: Hearing thresholds of 15 dB: do you mean that all had thresholds lower than 15 dB at all frequencies and at all repeated audiogram measurements?
We thank Reviewer #2 for spotting this error: we meant thresholds below 15dB HL. This has been corrected (L463). Indeed participants were submitted to several audiograms between fMRI sessions, to ensure no hearing loss could be caused by the scanner noise in these repeated sessions.
L472: were the 4 category levels balanced across the dataset (in number of occurrences of each category combination)?
The dataset was fully balanced, with an equal number of samples for each combination of language, gender, age, and identity. Furthermore, to minimize potential adaptation effects, the stimuli were also balanced within each run according to these categories, and identity was balanced across sessions. We made this clearer in Main voice stimuli (L492-L496).
L482: the test stimuli were selected as having high energy by the amplitude envelope. It is unclear what this means (how is the envelope extracted, what feature of it is used to measure 'high energy'?)
The selection of sounds with high energy was based on analyzing the amplitude envelope of each signal, which was extracted using the Hilbert transform and then filtered to refine the envelope. This envelope, which represents the signal's intensity over time, was used to measure the energy of each stimulus, and those that exceeded an arbitrary threshold were selected. From this pool of high-energy stimuli, likely including vowels, we selected six stimuli to be repeated during the scanning session, then reconstructed via decoding. This has been clarified in the text (L483-L484).
L500 was the audio filtered to account for the transfer function of the Sensimetrics headphones?
We did not perform any filtering, as the transfer function of the Sensimetrics is already very satisfactory as is. This has been clarified in the text (L503).
L500: what does 'comfortable level' correspond to and was it set per session (i.e. did it vary across sessions)?
By comfortable we mean around 85 dB SPL. The audio settings were kept similar across sessions. This has been added to the text (L504).
L526- does the normalization imply that the reconstructed spectrograms are normalized? Were the reconstructions then scaled to undo the normalization before inversion?
The paragraph on spectrogram standardization was not well placed inducing confusion. We have placed this paragraph in its more suitable location, in the Deep learning section (L545L550)
L606: does the identity GLM model the denoised betas from the first GLM or simply the BOLD data? The text indicates the latter, but I suspect the former.
Indeed: this has been clarified (L601-L602).
L704: could you unpack this a bit more? It is not easy to see why you specify the summing in the objective. Shouldn't this just be the ridge objective for a given voxel/ROI? Then you could just state it in matrix notation.
Thanks for pointing this out: we kept the formula unchanged but clarified the text, in particular specified that the voxel id is the ith index (L695).
L716: you used robust scaling for the classifications in latent space but haven't mentioned scaling here. Are we to assume that the same applies?
Indeed we also used robust scaling here, this is now made clear (L710-L711).
L720: Pearson correlation as a performance metric and its variance will depend on the choice of test/train split sizes. Can you show that the results generalize beyond your specific choices? Maybe the report explained variance as well to get a better idea of performance.
We used a standard 80/20 split. We think it is beyond the scope of this study to examine the different possible choices of splits, and prefer not to spend additional time on this point which we think is relatively minor.
Could you specify (somewhere) the stimulus timing in a run? ISI and stimulus duration are mentioned in different places, but it would be nice to have a summary of the temporal structure of runs.
This is now clarified at the beginning of the Methods section (L437-441)
Reviewer #3 (Recommendations For The Authors):
Code and data are not currently available.
Code and preprocessed data are now available (L826-827).
In the supplementary material, it would be beneficial to present the different analyses as boxplots, as in the main text, but with the ROIs in the left and right hemispheres separated, to better show potential hemispheric effect. Although this information is available in the Supplementary Tables, it is currently quite tedious to access it.
Although we provide the complete data split by hemisphere in the Tables, we do not believe it is relevant to illustrate left/right differences, as we do not have any hypotheses regarding hemispheric lateralization–and we would be underpowered in any case to test them with only three points by hemisphere.
In Figure 3a, it might be beneficial to order the identities by age for each gender in order to more clearly illustrate the structure of the RDMs,
The identities are indeed already ordered by increasing age: we now make this clear.
In Figure 3b, the variance for the correlations for the aTVA is higher than in other regions, why?
Please note that the error bar indicates variance across only 6 data points (3 subjects x 2 hemispheres) such that some fluctuations are to be expected.
Please make sure that all acronyms are defined, and that they are redefined in the figure legends.
This has been done.
Gender and age are primarily encoded by different brain regions (Figure 5, pTVA vs aTVA). How does this finding compare with existing literature?
This interesting finding was not expected. The cerebral processing of voice gender has been investigated by several groups including ours (Charest et al., 2013, Cerebral Cortex). Using an fMRI-adaptation design optimized using a continuous carry-over protocol and voice gender continua generated by morphing, we found that regions dealing with acoustical differences between voices of varying gender largely overlapped with the TVAs, without clear differentiation between the different subparts. Evidence for the role of the different TVAs in voice age processing remains scarce.
-
-
www.biorxiv.org www.biorxiv.org
-
Reviewer #1 (Public review):
Summary:
This study investigated the immunogenicity of a novel bivalent EABR mRNA vaccine for SARS-CoV-2 that expresses enveloped virus-like particles in pre-immune mice as a model for boosting the population that is already pre-immune to SARS-CoV-2. The study builds on promising data showing a monovalent EABR mRNA vaccine induced substantially higher antibody responses than a standard S mRNA vaccine in naïve mice. In pre-immune mice, the EABR booster increased the breadth and magnitude of the antibody response, but the effects were modest and often not statistically significant.
Strengths:
Evaluating a novel SARS-CoV-2 vaccine that was substantially superior in naive mice in pre-immune mice as a model for its potential in the pre-immune population.
Weaknesses:
(1) Overall, immune responses against Omicron variants were substantially lower than against the ancestral Wu-1 strain that the mice were primed with. The authors speculate this is evidence of immune imprinting, but don't have the appropriate controls (mice immunized 3 times with just the bivalent EABR vaccine) to discern this. Without this control, it's not clear if the lower immune responses to Omicron are due to immune imprinting (or original antigenic sin) or because the Omicron S immunogen is just inherently more poorly immunogenic than the S protein from the ancestral Wu-1 strain.
(2) The authors reported a statistically significant increase in antibody responses with the bivalent EABR vaccine booster when compared to the monovalent S mRNA vaccine, but consistently failed to show significantly higher responses when compared to the bivalent S mRNA vaccine, suggesting that in pre-immune mice, the EABR vaccine has no apparent advantage over the bivalent S mRNA vaccine which is the current standard. There were, however, some trends indicating the group sizes were insufficiently powered to see a difference. This is mostly glossed over throughout the manuscript. The discussion section needs to better acknowledge these limitations of their studies and the limited benefits of the EABR strategy in pre-immune mice vs the standard bivalent mRNA vaccine.
(3) The discussion would benefit from additional explanation about why they think the EABR S mRNA vaccine was substantially superior in naïve mice vs the standard S mRNA vaccine in their previously published work, but here, there is not much difference in pre-immune mice.
-
-
social-media-ethics-automation.github.io social-media-ethics-automation.github.io
-
One useful way to think about harassment is that it is often a pattern of behavior that exploits the distinction between things that are legally proscribed and things that are hurtful, but not so harmful as to be explicitly prohibit by law given the protection of freedoms. Let’s use an example to clarify.
This chapter make me see harassment very different than before. I used to think it is only about big, obvious things like death threats, but the puddle example shows how many small actions together can still really hurt someone. Online it’s even worse, because people can pretend every single comment is “not that serious” while the target already feel scared and tired. I still don’t fully know where the line should be between free speech and moderation, but now it’s harder to say “it’s just the internet, just ignore it,” because clearly it’s not that simple.
-